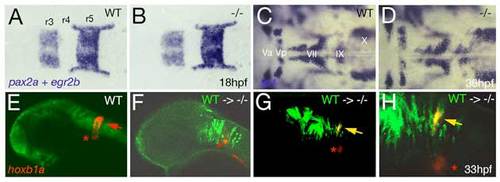
Loss of Brpf1 function in the hindbrain has a cell-autonomous effect on hoxb1a expression, but no obvious effects on anterior-posterior patterning. (A-D) Hindbrain patterning of brpf1 mutants appears normal. (A,B) egr2b (krox20) (Oxtoby and Jowett, 1993) and pax2a (Krauss et al., 1991) expression at 18 hpf, dorsal view of hindbrain region. In the brpf1 mutant (B; genotyped after photography), the width of rhombomeres r3, r4 and r5 is similar to that in wild-type siblings (A) (n=13/13). There was some variability (see r3), which, however, did not correlate with the brpf1 genotype. This is in contrast to the shifts obtained upon MO-based knockdown of hoxb1 genes, with a broadening of r3, whereas r4 and r5 are narrower than in uninjected siblings (McClintock et al., 2002). (C,D) In situ hybridization for isl1 transcripts (Inoue et al., 1994), 36 hpf, dorsal view of hindbrain region. Branchiomotor neurons of the trigeminal (V), facial (VII), glossopharyngeal (IX) and vagal nerves (X) are indicated. During normal development, cell bodies of the Vth nerve differentiate in r2 (Va) and r3 (Vp), where they remain during further development. Cell bodies of the VIIth nerve differentiate in rhombomers r4 and r5, followed by posterior migration to end up in r6 and r7 at 36 hpf (McClintock et al., 2002). brpf1 mutant (D; genotyped after photography) (n=21/12) and wild-type sibling show no significant difference in the patterning of isl1-positive branchiomotor cell bodies. The same results were obtained for mutants at 24 hpf, 28 hpf, 32 hpf and 40 hpf. This is in contrast to the phenotype observed in hoxb1 morphants, in which cell bodies of the Vth nerve remain in lateral positions of r4/5 (McClintock et al., 2002). Also in moz mutants, no defects during early rhomobomere patterning could be detected, while branchiomotor defects were much weaker and less penetrant than in hoxb1 morphants (Miller et al., 2004). Consistently, we found that Hox gene expression in the hindbrain of brpf1 mutants was much less affected than in the cranial neural crest (CNC) (see Fig. 2E-H and Fig. S2E,F). Together, this suggests that in the hindbrain, Brpf1/Moz activity and/or maintenance of anterior Hox gene expression is less critical for segmental identity determination than in the CNC. (E-H) Brpf1 promotes hoxb1a expression in hindbrain r4 cells in a cell-autonomous fashion. Fluorescein-dextran (Molecular Probes) was injected into wild-type donor embryos at the 1- to 4-cell stage. For hindbrain transplants, labeled cells were taken from the respective region (Kimmel et al., 1990) of shield stage (6 hpf) donor embryos and homotopically transferred into unlabeled shield stage brpf1 morphant hosts. Chimeric embryos were fixed at 35 hpf and in situ hybridized for hoxb1a, using Fast Red (Roche) as substrate. Subsequently, transplanted cells were stained by anti-fluorescein immunostaining. Chimeric embryos were analyzed by confocal laser-scanning microscopy (Zeiss LSM510 META). Lateral views of heads of wild-type (E) or brpf1 morphant embryos with transplanted wild-type cells (green; F-H). (H) A higher magnification of G. hoxb1a expression (in red) in ventral medial nucleus, which serves as internal control, is indicated by red asterisks. Chimeric embryos with wild-type cells anterior and posterior to r4 lacked hoxb1a expression (F; n=12/12), whereas wild-type cells within r4 were strongly hoxb1a-positive (H; yellow arrows; n=4/4). Note that hoxb1a expression remains absent in brpf1 morphant cells adjacent to hoxb1a-positive wild-type cells, ruling out the possibility that Brpf1 acts via a Hox expression-activating extracellular signal such as retinoic acid, which has a posteriorizing effect on Hox gene expression and segmental identity (Glover et al., 2006).
|

