- Title
-
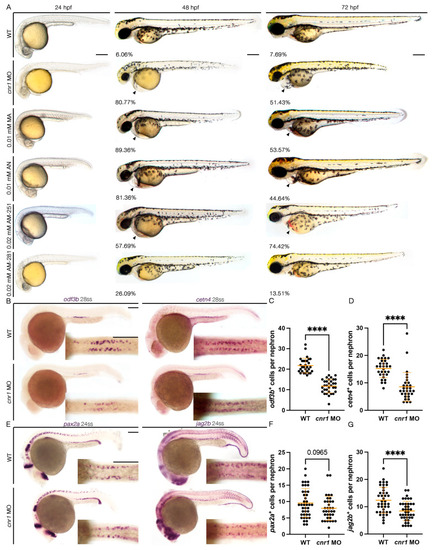
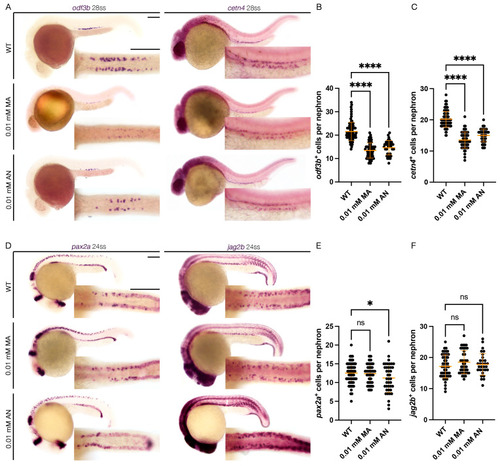
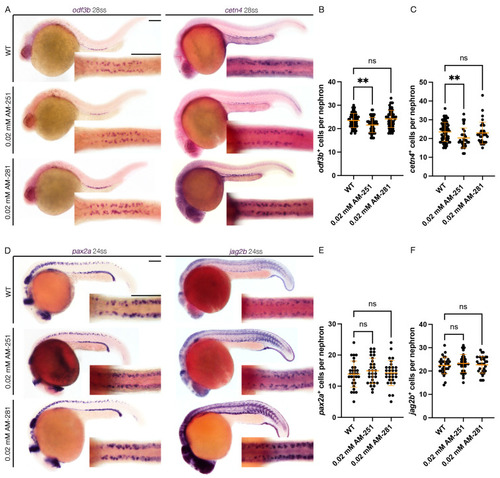
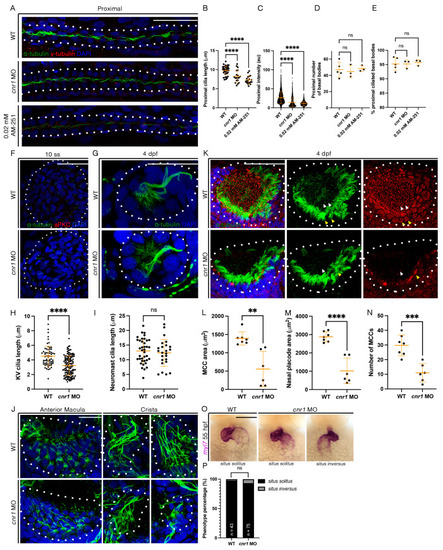
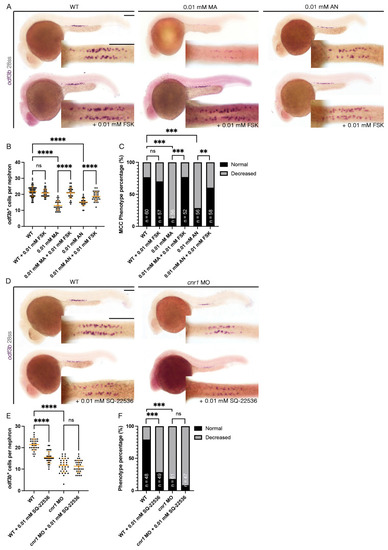
Cannabinoid Receptor 1 Regulates Zebrafish Renal Multiciliated Cell Development via cAMP Signaling
- Authors
- Nguyen, T.K., Baker, S., Angtuaco, J., Arceri, L., Kaczor, S., Fitzsimonds, B., Hawkins, M.R., Wingert, R.A.
- Source
- Full text @ J Dev Biol
|
|
|
|
|
|
|
|
|
|
|
Roadmap delineating the relationship between Cnr1 and players in renal MCC development. |