- Title
-
Restoration of retinal regenerative potential of Müller glia by disrupting intercellular Prox1 transfer
- Authors
- Lee, E.J., Kim, M., Park, S., Shim, J.H., Cho, H.J., Park, J.A., Park, K., Lee, D., Kim, J.H., Jeong, H., Matsuzaki, F., Kim, S.Y., Kim, J., Yang, H., Lee, J.S., Kim, J.W.
- Source
- Full text @ Nat. Commun.
|
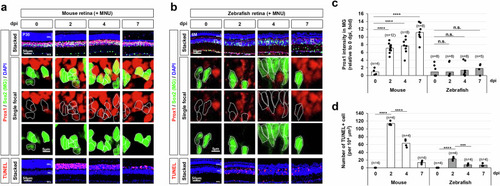
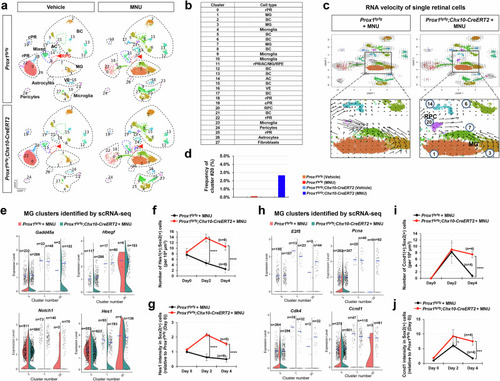
Injury-induced accumulation of Prox1 in mouse MG. |
|
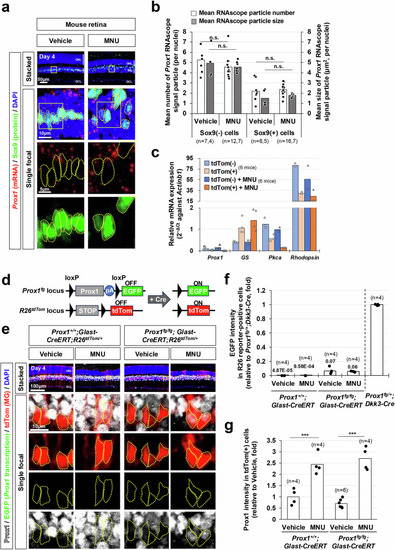
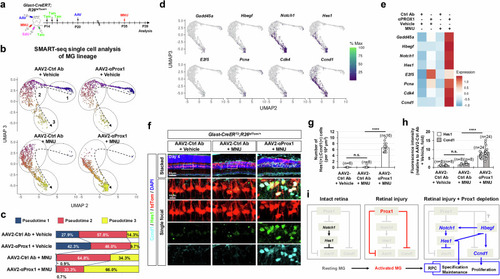
Exogenous origin of Prox1 in mouse MG. |
|
|
|
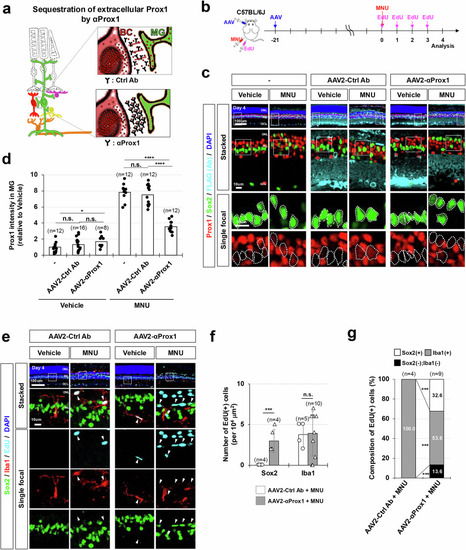
Sequestration of extracellular Prox1 by anti-Prox1 antibody restores MG proliferative potential. |
|
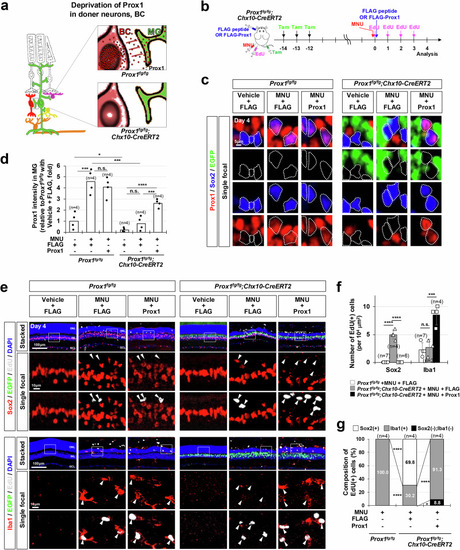
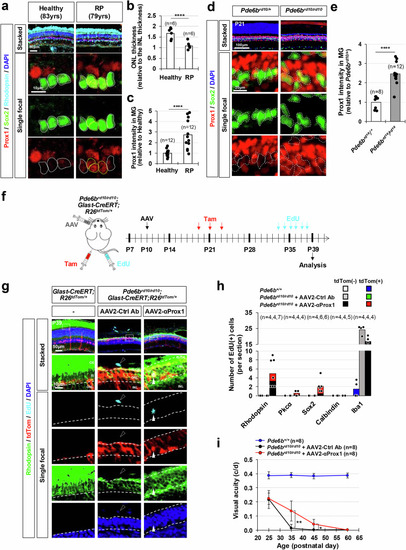
Injury-induced emergence of RPCs in the mouse retina following Prox1 deletion in BCs. |
|
Injury-induced reprogramming of MG into RPCs upon reduced Prox1 transfer. |
|
Delaying vision loss in early-onset RP model mice via viral gene delivery of anti-Prox1 antibody. |
|
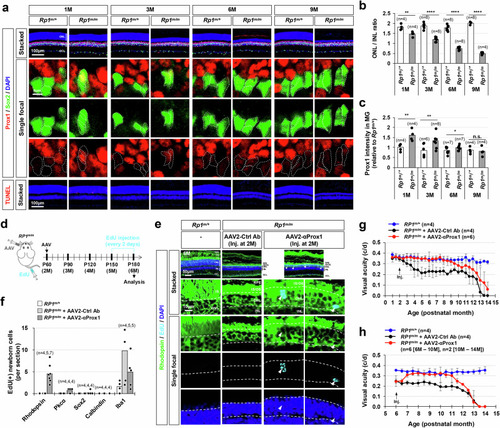
Vision recovery in late-onset RP model mice by viral gene delivery of anti-Prox1 antibody. ( |