- Title
-
Creation of a novel zebrafish model with low DHA status to study the role of maternal nutrition during neurodevelopment
- Authors
- Ranard, K.M., Appel, B.
- Source
- Full text @ J. Lipid Res.
|
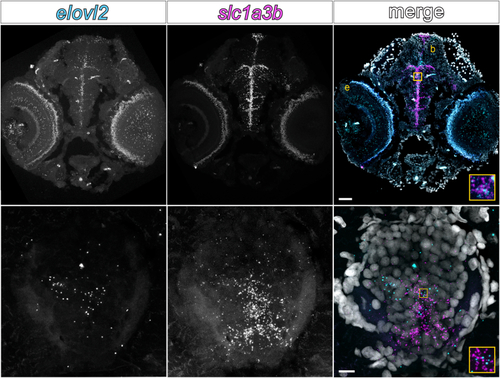
Zebrafish central nervous system tissues express EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Wild-type zebrafish fed a DHA-deficient diet have similar growth trajectories as fish fed DHA-sufficient diet. A: Average fish body mass, calculated by dividing total tank wet mass by the number of fish per tank. B: Average fish length (nose to caudal peduncle), calibrated to ruler in tank. Separate cohorts of fish were fed the assigned diets starting at either 30 or 60 dpf, indicated in the plots by square or circle symbols, respectively. The main effects of diet and time (weeks on diet) were analyzed by 2-way repeated measures ANOVAs for the 30 and 60 dpf cohorts separately. Body mass and length did not differ between the SUFF and DEF groups for either initiation age. As expected for developing zebrafish, body mass and length significantly increased with feeding duration (body mass: 30 dpf, PHENOTYPE:
|
|
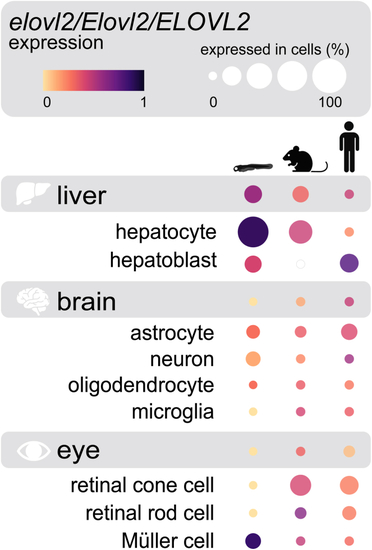
Zebrafish, mouse, and human central nervous system tissues express |
|
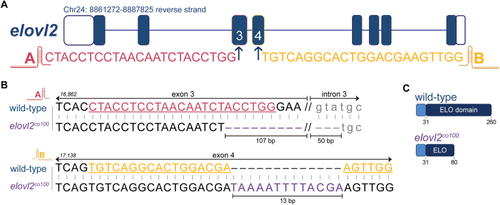
Generation of |
|
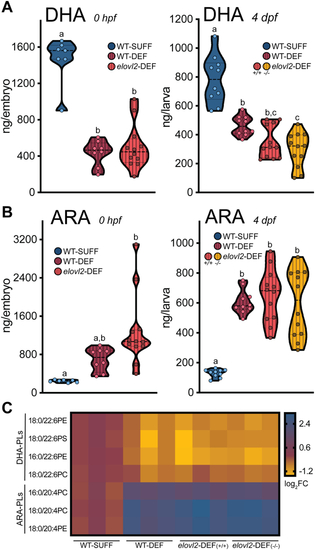
Maternal dietary DHA restriction and biosynthesis disruption depletes offspring DHA and increases ARA. Adult wild-type and |
|
DHA-restricted mothers produce offspring with a series of morphological phenotypes, and phenotype severity is related to offspring DHA and ARA status. A: Example 4 dpf larvae from each experimental group. The PHENOTYPE:
|
|
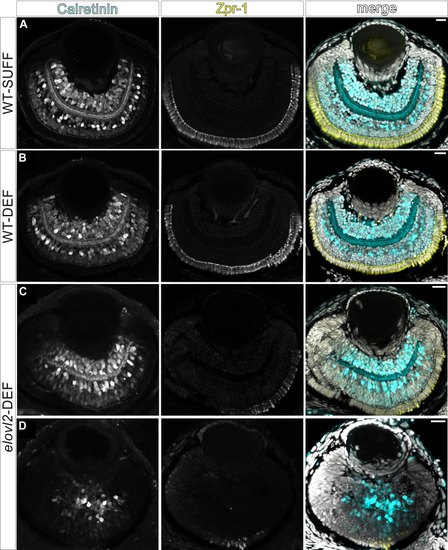
DHA-deficient |
|
Offspring with low DHA status have dysregulated vision and stress response gene pathways. Morphologically normal 4 dpf larvae from wild-type mothers fed a DHA-sufficient diet (WT-SUFF) or |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|