- Title
-
Image restoration of degraded time-lapse microscopy data mediated by near-infrared imaging
- Authors
- Gritti, N., Power, R.M., Graves, A., Huisken, J.
- Source
- Full text @ Nat. Methods
|
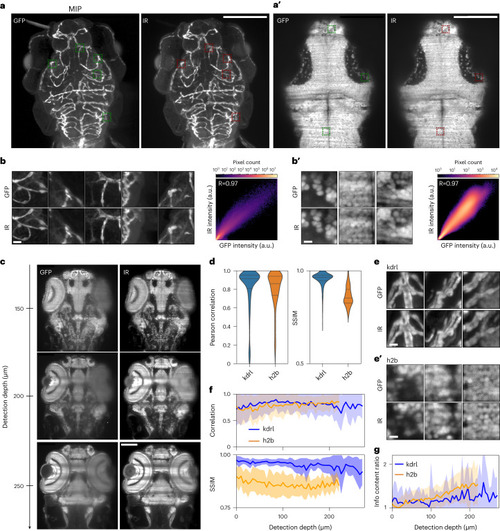
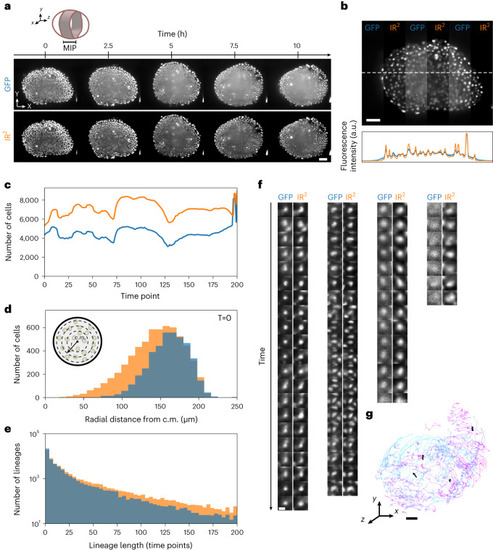
Highly selective near-infrared staining and light-sheet microscopy affords superior imaging at depth in tissue. |
|
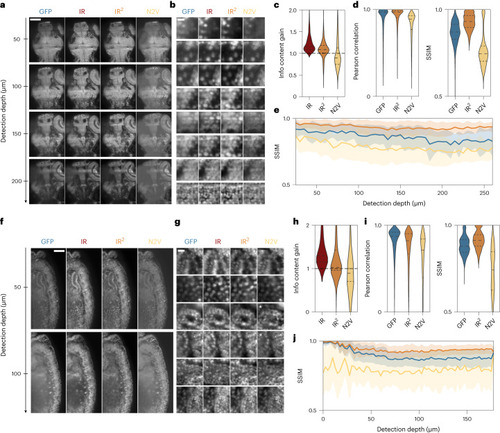
Infrared-mediated image restoration improves image quality of degraded GFP images. |
|
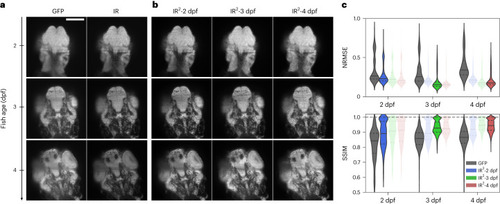
The quality of IR2 images is robust over large developmental intervals. |
|
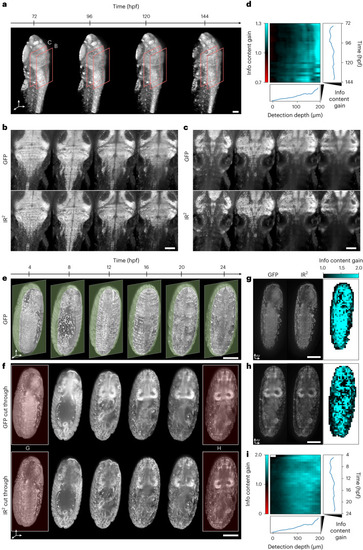
Infrared-mediated image restoration provides high-contrast deep-tissue time-lapse imaging of living biological systems. |
|
Image restoration of developing pescoid. |