- Title
-
RhoA GEF Mcf2lb regulates rosette integrity during collective cell migration
- Authors
- Olson, H.M., Maxfield, A., Calistri, N.L., Heiser, L.M., Qian, W., Knaut, H., Nechiporuk, A.V.
- Source
- Full text @ Development
|
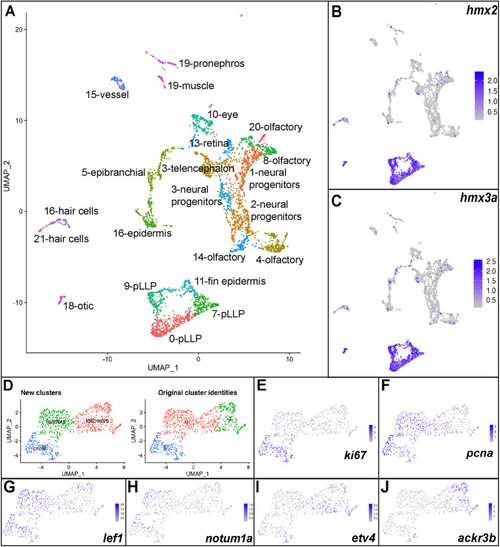
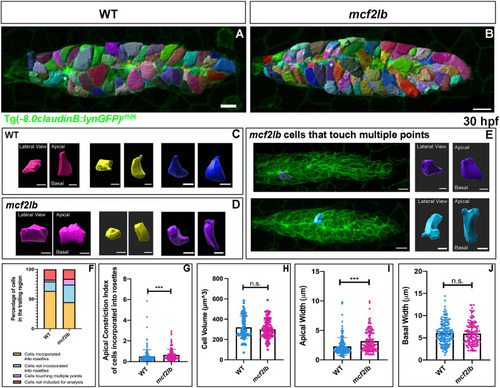
EXPRESSION / LABELING:
|
|
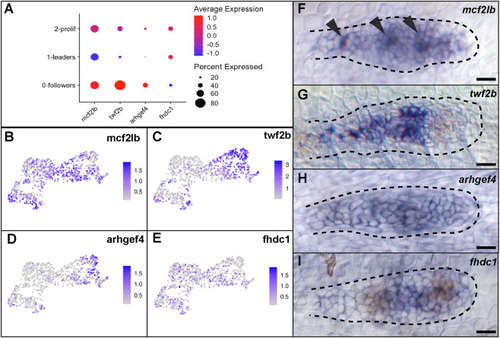
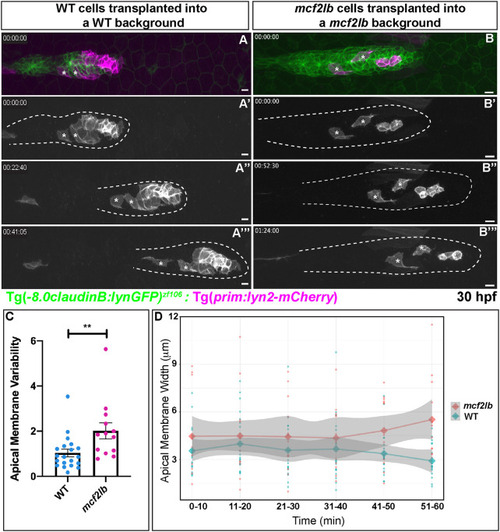
EXPRESSION / LABELING:
|
|
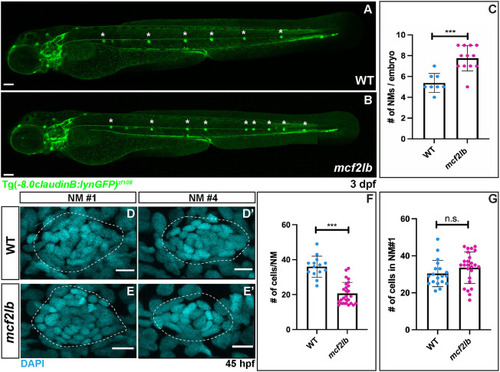
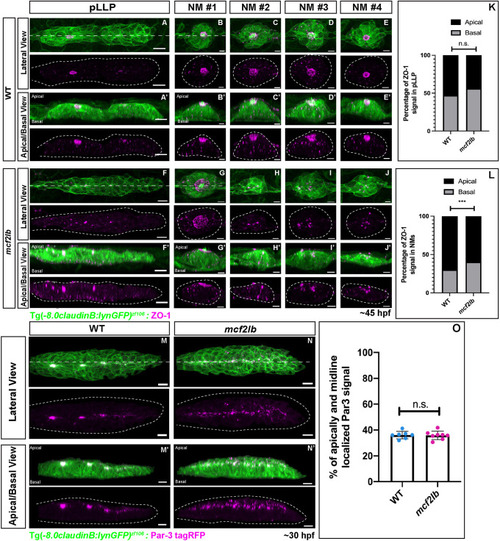
EXPRESSION / LABELING:
PHENOTYPE:
|
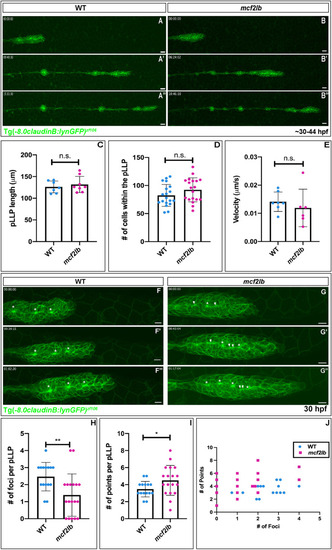
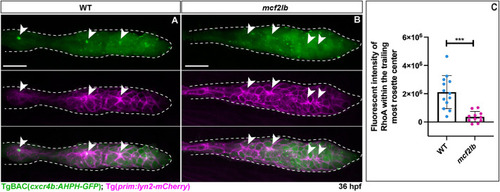
|
|
|
|
|
|
|
|
|
|
|
|