- Title
-
Foxn1 is not essential for T-cell development in teleosts
- Authors
- Schorpp, M., Swann, J.B., Hess, I., Ho, H.C., Pietsch, T.W., Boehm, T.
- Source
- Full text @ Eur. J. Immunol.
|
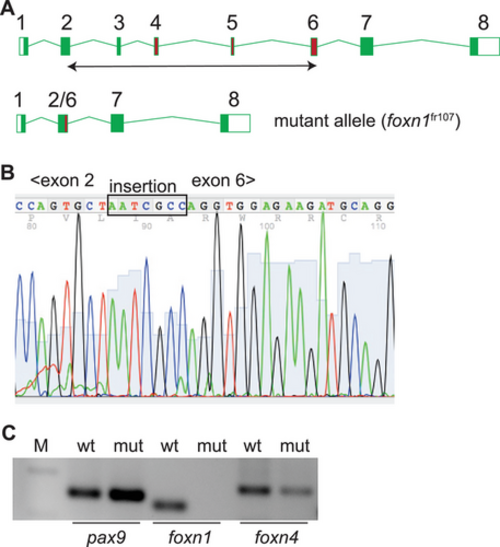
Characterization of a foxn1 null allele in zebrafish. (A) Schematic structure of the zebrafish foxn1 gene; exons are numbered, and the exons encoding the DNA binding domain are indicated as red rectangles. A double-headed arrow below the schematic indicates the region deleted in the foxn1fr107 mutant allele. (B) Partial sequence of the cDNA spanning the deletion from within exon 2 to within exon 6; note the insertion of seven nucleotides as a result of the CRISPR/Cas9-mediated rearrangement that fuses parts of exons 2 and 6. (C) RT-PCR analysis of the indicated epithelial genes in the thymi of wildtype (wt) and foxn1-deficient (mut) zebrafish (M; marker lane; amplicon sizes: pax9, 149 bp; foxn1, 110 bp; foxn4, 174 bp. Note the absence of an intact foxn1 transcript in the mutant thymus. EXPRESSION / LABELING:
PHENOTYPE:
|
|
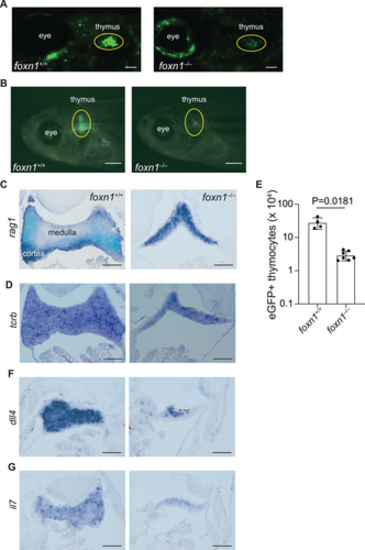
Impaired intrathymic lymphopoiesis in foxn1-deficient fish. (A) Whole mount depiction of zebrafish at 6 dpf, transgenic for an ikzf1:eGFP pan-lymphocyte reporter transgene. The foxn1 genotypes are indicated, as are anatomical landmarks; note that subpopulations of neurons also express ikzf1. Scale = 0.1 mm. (B) Macroscopic appearance of adult fish (6 weeks of age) transgenic for the ikzf1:eGFP reporter transgene. The foxn1 genotypes are indicated. Scale = 1 mm. (C) RNA in situ hybridization of thymic tissue sections using a rag1-specific probe. Note that the medulla (rag1-negative region) is barely visible in the mutants. (D) RNA in situ hybridization of thymic tissue sections using a tcrb-specific probe. (E) Adult mutant fish (6 weeks of age) possess fewer eGFP-positive thymocytes as determined by flow cytometry on cells isolated from microdissected thymic lobes (mean ± s.e.m. are shown; t-test, two-tailed). Each data point represents an individual fish. (F) RNA in situ hybridization of thymic tissue sections using a dll4-specific probe. (G) RNA in situ hybridization of thymic tissue sections using an il7-specific probe. For C, D, F, G, scale, 0.1 mm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
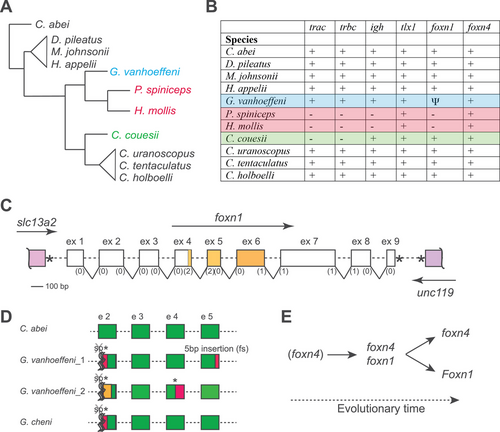
Characteristics of immunogenomes of selected Ceratioidei. (A) Schematic cladogram of phylogenetic relationships among the indicated species (data taken from the study by Swann et al. [24]). (B) Gene content in the indicated species. +, gene present and intact; –, gene missing; Ψ, pseudogenized gene. Further information on tlx1, foxn4, and foxn1 is presented in Supporting information Figs. S2–S4, and Supporting information Data S1–S3. (C) Schematic structure of the foxn1 gene and its syntenic relationship; the exons (but not introns) are drawn to scale with phases of splice sites indicated in brackets. The slc13a2 gene has the same transcriptional orientation as foxn1, whereas the unc119 gene is transcribed in the opposite direction. The * denotes the stop codons. (D) Schematic of mutations observed in foxn1 genes of the indicated specimens (see Supporting information Figs. S5–S7 for details). The splice acceptor sites of exon 2 are mutated, which suggests the presence of aberrant splicing patterns (symbolized by crossed sp, *, and wiggly lines). Specimen G. vanhoeffeni_1 corresponds to specimen UW118701 described in the study by Swann et al. [24]. Exonic sequences shaded in red or orange indicate regions different from wild-type sequences. (E) Scheme highlighting the evolutionary sequence of activities of Foxn1/4 transcription factor-family members supporting thymopoiesis during vertebrate evolution. Primordial thymopoietic activity may have been supported by Foxn4 alone; however, after gene duplication leading to the emergence of Foxn1, co-expression of the two thymopoietic factors established a robust genetic network. This robustness was subsequently abolished in two ways, by loss of an intact foxn1 gene in some species of deep-sea anglerfishes (resulting in foxn4 activity only), and by loss of expression of Foxn4 in the thymic epithelium of mammals (resulting in Foxn1 activity only). |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|