- Title
-
Activator-blocker model of transcriptional regulation by pioneer-like factors
- Authors
- Riesle, A.J., Gao, M., Rosenblatt, M., Hermes, J., Hass, H., Gebhard, A., Veil, M., Grüning, B., Timmer, J., Onichtchouk, D.
- Source
- Full text @ Nat. Commun.
|
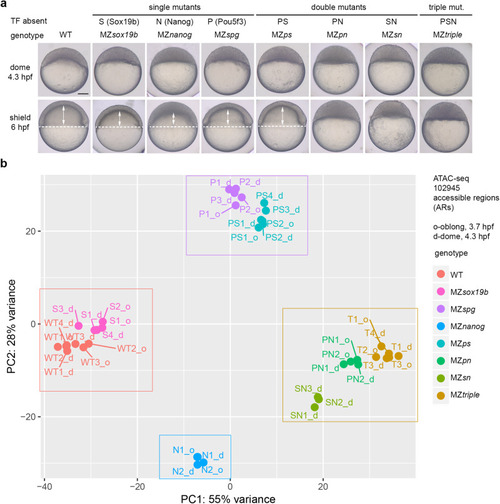
Single and combined mutants by zebrafish zygotic genome activators used in this study. |
|
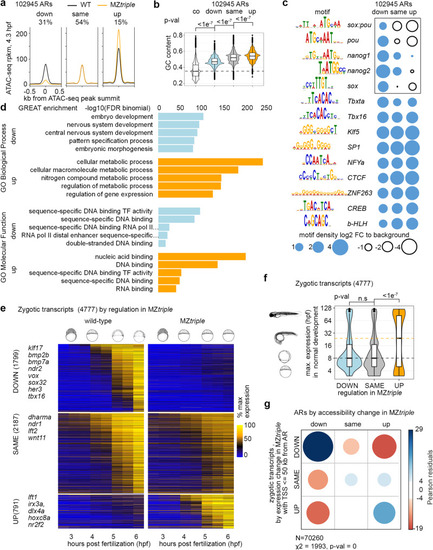
Changes in chromatin accessibility parallel the changes in gene expression in Pou5f3, Sox19b and Nanog triple mutant. |
|
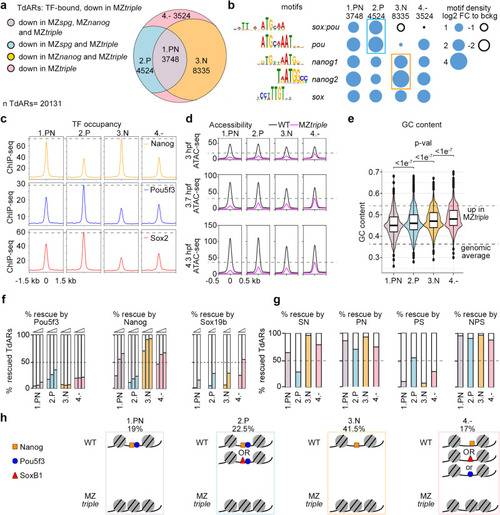
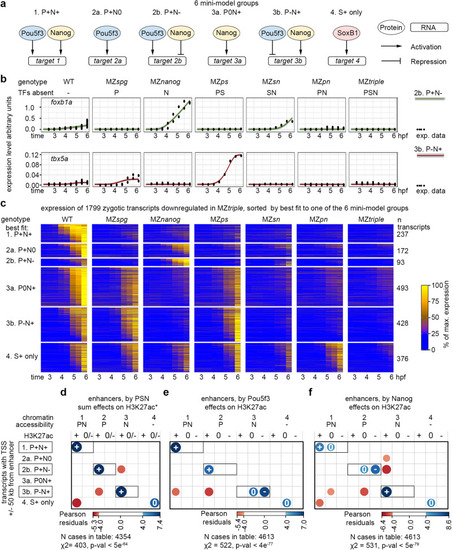
Four types of TdARs by pioneer-like activity of Pou5f3, SoxB1, and Nanog. |
|
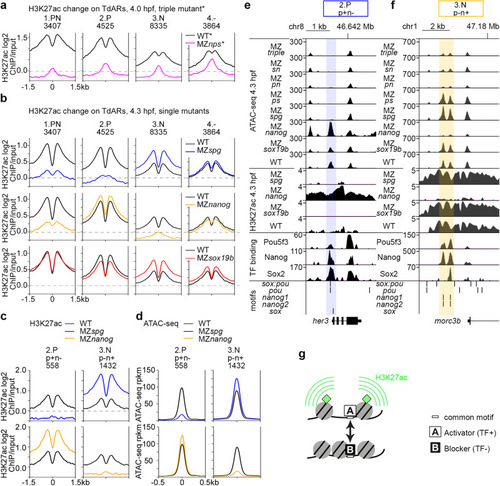
Activator-blocker model: Pou5f3 and Nanog oppose each other effects on H3K27 acetylation and chromatin accessibility on a fraction of enhancers. |
|
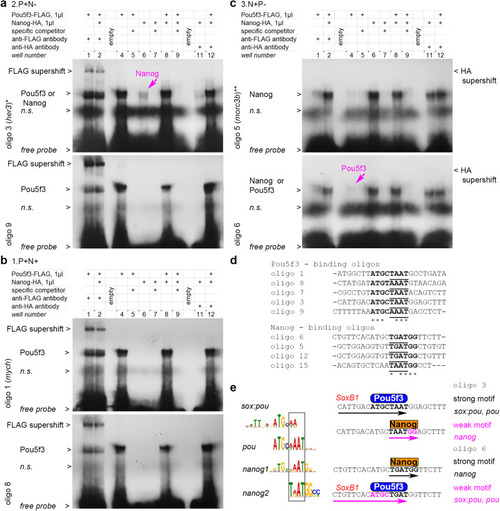
Pou5f3 and Nanog bind in a mutually exclusive manner to overlapping motifs. |
|
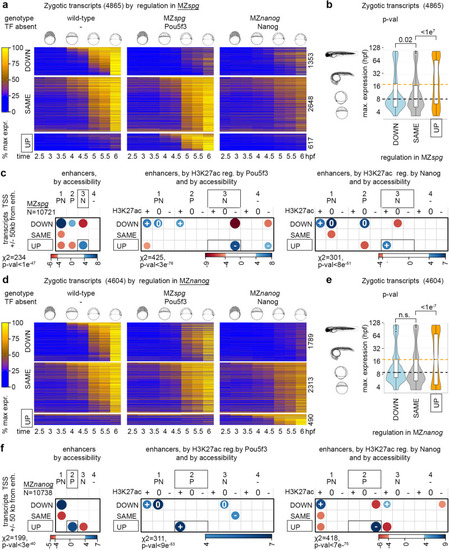
Zygotic gene expression is balanced by synergy and competition of Pou5f3 and Nanog on common enhancers. |
|
Pou5f3 blocks premature transcriptional activation by Nanog and vice versa. |
|
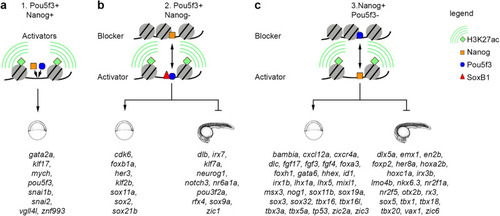
Synergistic and antagonistic Pou5f3/Nanog direct enhancers: the mechanism of action and direct target genes. |