- Title
-
The mesencephalic locomotor region recruits V2a reticulospinal neurons to drive forward locomotion in larval zebrafish
- Authors
- Carbo-Tano, M., Lapoix, M., Jia, X., Thouvenin, O., Pascucci, M., Auclair, F., Quan, F.B., Albadri, S., Aguda, V., Farouj, Y., Hillman, E.M.C., Portugues, R., Del Bene, F., Thiele, T.R., Dubuc, R., Wyart, C.
- Source
- Full text @ Nat. Neurosci.
|
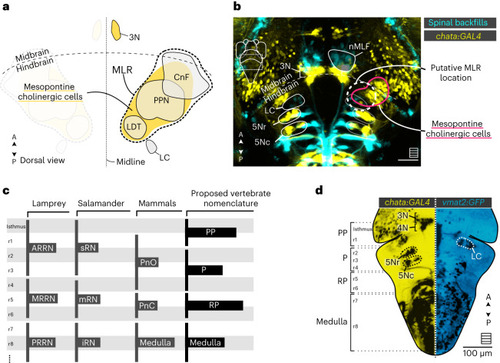
The putative location of the MLR in zebrafish inferred from previous studies in other vertebrates. |
|
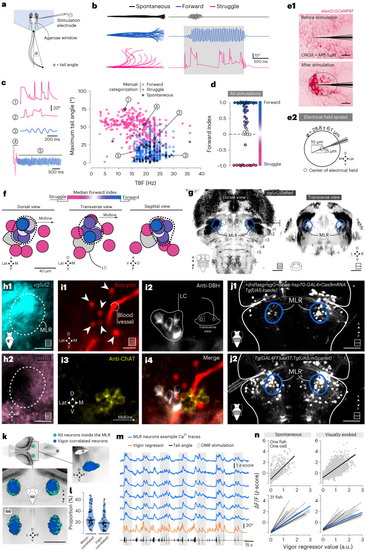
Anatomical and functional description of the MLR in larval zebrafish reveals neurons triggering forward locomotion and encoding vigor. |
|
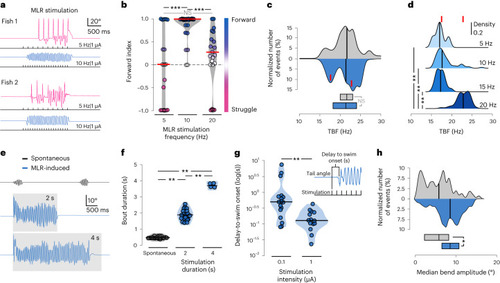
MLR stimulation parameters set the duration, time onset, amplitude and locomotor frequency of forward swimming |
|
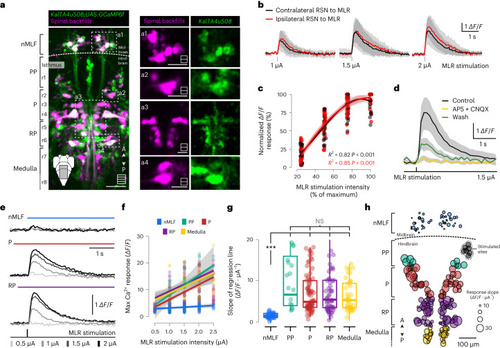
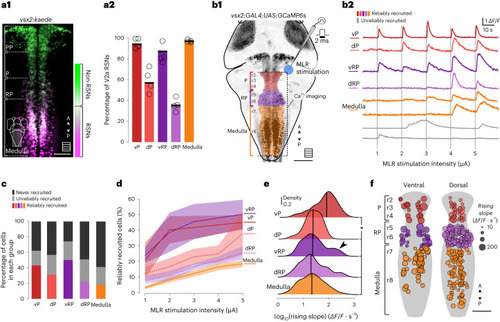
Unilateral MLR stimulation recruits ipsilateral and contralateral hindbrain RSNs but not the nMLF. |
|
Single-pulse MLR stimulations gradually recruit a subset of V2a RSNs. |
|
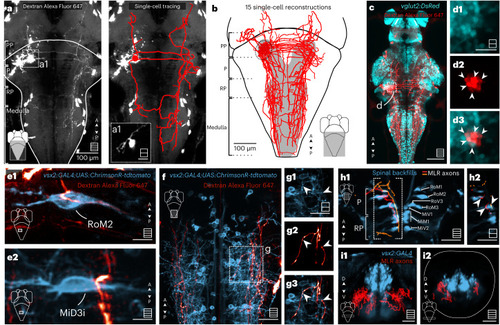
MLR neurons project bilaterally to V2a RSNs forming putative axosomatic synapses in the P and RP areas and axodendritic synapses in the medulla. |
|
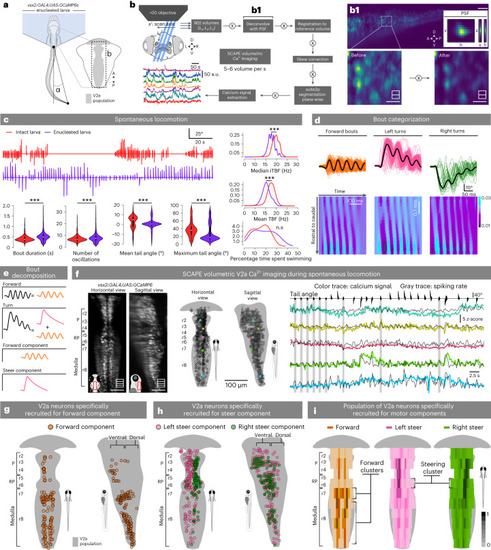
Whole-population brain stem V2a neurons recruitment during spontaneous swimming using high-speed, single-objective light sheet microscopy. |
|
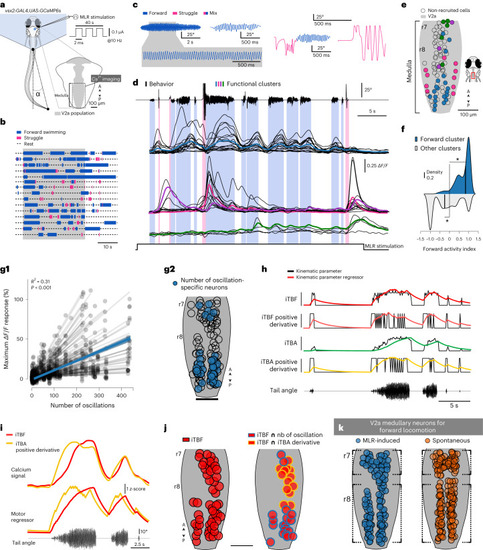
Medullary V2a RSNs are specifically recruited during MLR-induced forward swimming and encode the number of oscillations, TBF and amplitude. |