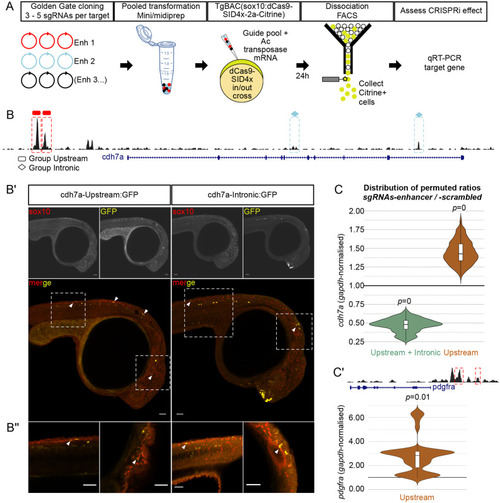
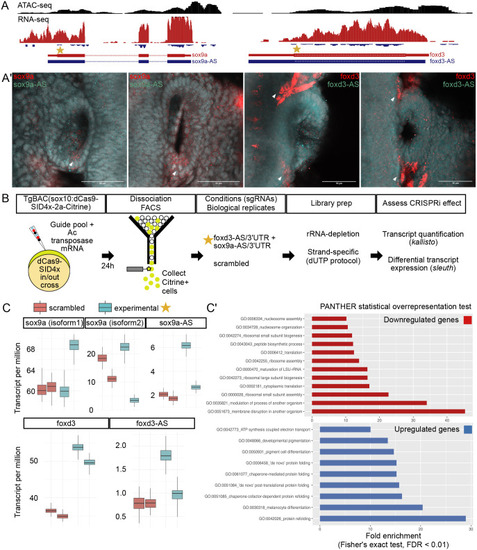
- Title
-
Ac/Ds transposition for CRISPR/dCas9-SID4x epigenome modulation in zebrafish
- Authors
- Chong-Morrison, V., Mayes, S., Simões, F.C., Senanayake, U., Carroll, D.S., Riley, P.R., Wilson, S.W., Sauka-Spengler, T.
- Source
- Full text @ Biol. Open
|
|
|
|
|
|
|
|