- Title
-
NERNST: a genetically-encoded ratiometric non-destructive sensing tool to estimate NADP(H) redox status in bacterial, plant and animal systems
- Authors
- Molinari, P.E., Krapp, A.R., Weiner, A., Beyer, H.M., Kondadi, A.K., Blomeier, T., López, M., Bustos-Sanmamed, P., Tevere, E., Weber, W., Reichert, A.S., Calcaterra, N.B., Beller, M., Carrillo, N., Zurbriggen, M.D.
- Source
- Full text @ Nat. Commun.
|
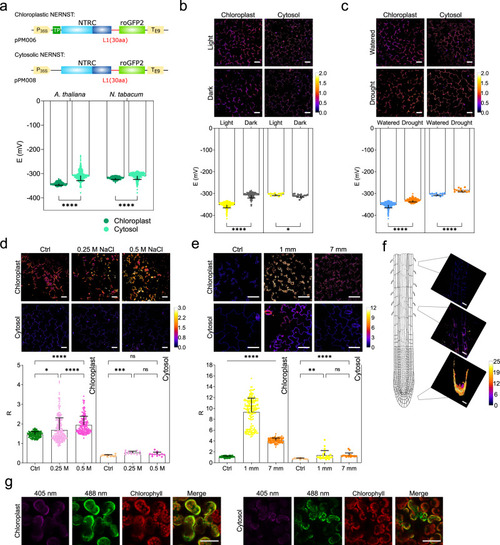
Design, construction and in vitro characterization of NERNST. |
|
NADP(H) redox state in |
|
In vivo analysis of the NADP(H) redox state in the cytosol and chloroplasts of leaf cells and roots. |
|
NERNST probes the NADP(H) redox status in isolated leaf protoplasts. Expression vectors used are those described in Fig. |
|
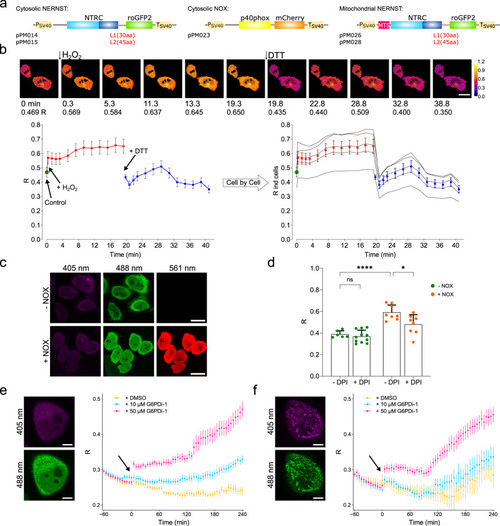
Mid-throughput analysis of NADP(H) dynamics in mammalian cells and mitochondria. |
|
Simultaneous imaging of NADP(H) and peroxide dynamics in a wounded region of zebrafish tail fin |