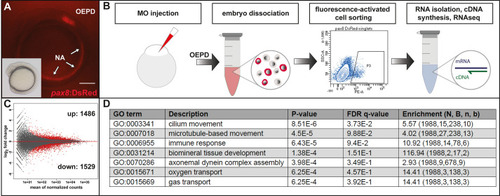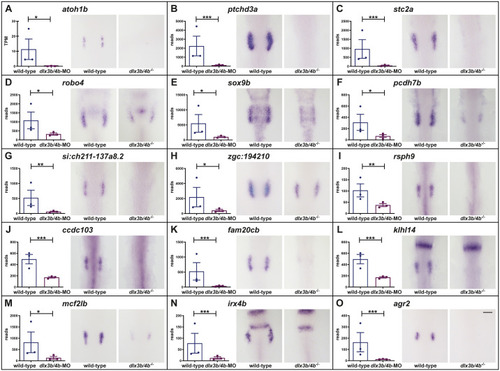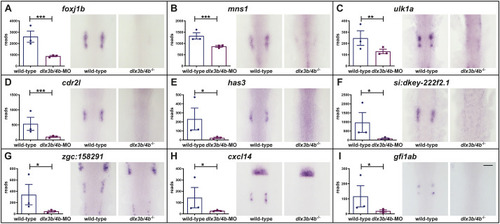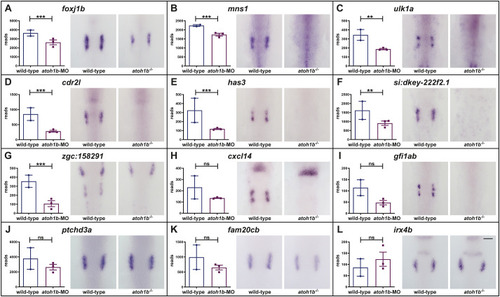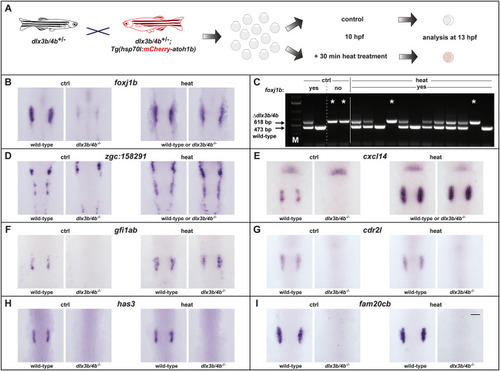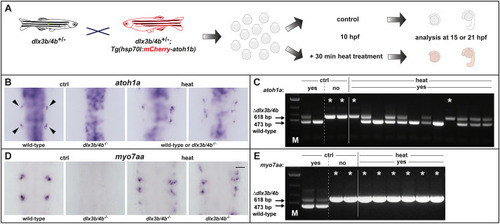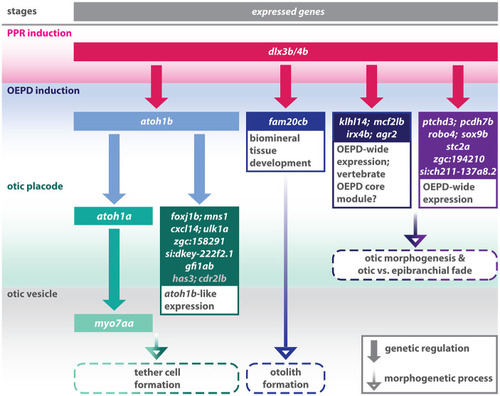
Schematic summary of the genetic events controlled by Dlx3b/4b during early otic development in zebrafish. Dlx3b/4b are expressed already during formation of the preplacodal region (PPR). After induction of the otic-epibranchial progenitor domain (OEPD), Dlx3b/4b control the onset of various genes which might govern various morphogenetic processes at subsequent placodal and vesicles stages. Among the genes showing an OEPD-wide expression (ptchd3, pcdh7b, si:ch211-137a8.2, robo4, sox9b, stc2a and zgc:194210), we also find klhl14; mcf2lb; irx4b and agr2 which have been reported to be expressed within the chicken OEPD (Chen et al., 2017). Hence, they might be members of a conserved, vertebrate core module regulating subsequent otic morphogenesis as well as otic versus epibranchial fate decisions. In addition, we find foxj1b, mns1, ulk1a, cxcl14, zgc:158291, si:dkey-222f2.1, gfi1ab, has3 and cdr2l, which display an atoh1b-like expression and might play a role in tether cell formation. Except has3 and cdr2l (grey), all genes of this module can be activated via ectopic expression of Atoh1b, even in the absence of Dlx3b/4b. Finally, we find that Dlx3b/4b controls the expression of fam20cb a gene associated with biomineral tissue development which might be important for subsequent otolith formation.
|