- Title
-
EmbryoNet: using deep learning to link embryonic phenotypes to signaling pathways
- Authors
- Čapek, D., Safroshkin, M., Morales-Navarrete, H., Toulany, N., Arutyunov, G., Kurzbach, A., Bihler, J., Hagauer, J., Kick, S., Jones, F., Jordan, B., Müller, P.
- Source
- Full text @ Nat. Methods
|
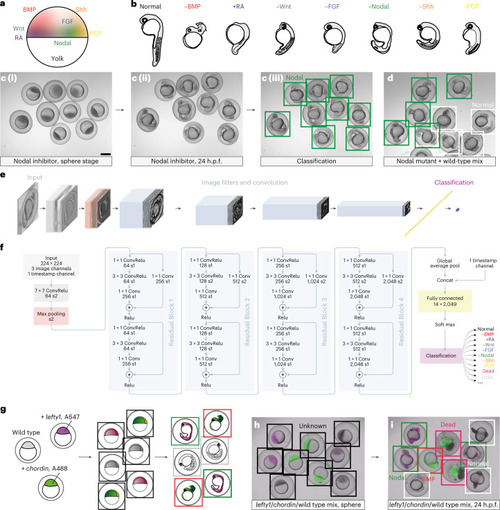
The CNN EmbryoNet robustly identifies molecular defects based on phenotype data. |
|
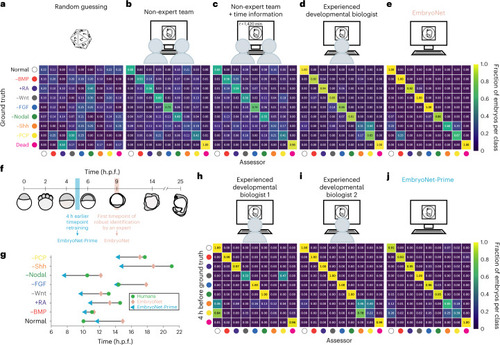
Classification of 98 single embryo images by non-expert teams, experienced researchers and EmbryoNet |
|
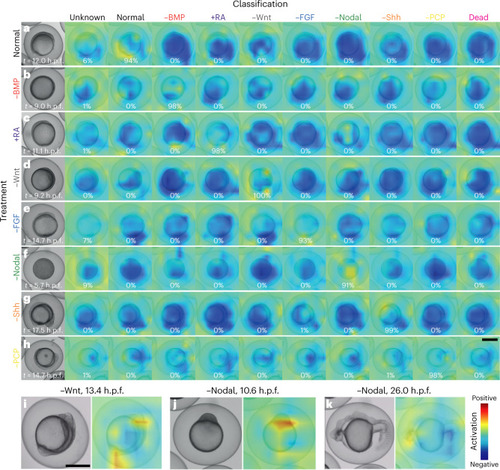
Embryo features activating the neural network. Class activation heatmaps based on the last convolutional layer of EmbryoNet-Prime showing the part of the image that activates the network at the given timepoint for normal ( |
|
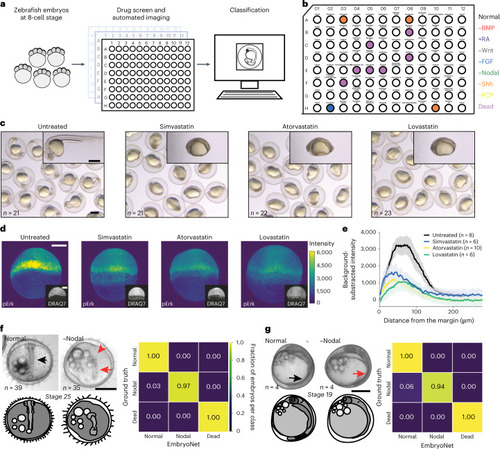
Applications of EmbryoNet in drug screening and other species. |