- Title
-
A p21-GFP zebrafish model of senescence for rapid testing of senolytics in vivo
- Authors
- Morsli, S., Henriques, C.M., Ellis, P.S., Mortiboys, H., Baxendale, S., Loynes, C.A., Renshaw, S.A., Bellantuono, I.
- Source
- Full text @ Aging Cell
|
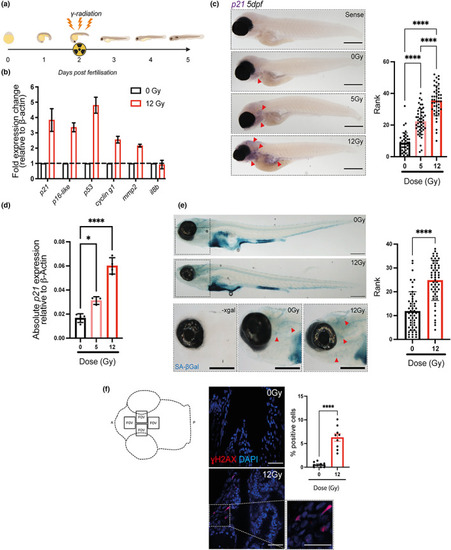
Irradiation of zebrafish larvae upregulates multiple markers of senescence. (a) Diagram depicting the experimental protocol used to induce senescence in zebrafish larvae using Cs137 ɣ‐Irradiation at 2 dpf and assessing markers of senescence at 5 dpf. (b) Quantitative PCR (qPCR) of whole zebrafish mRNA at 5 dpf following 12 Gy irradiation to determine gene expression of |
|
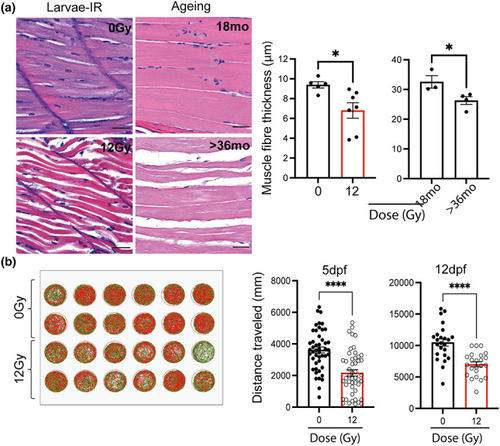
Zebrafish larvae showed reduced muscle fibre thickness and mobility following irradiation. (a) Representative photomicrographs of haematoxylin and eosin (H&E) staining of zebrafish muscle from either 12 dpf larvae with and without irradiation (top left panel), and middle aged and geriatric adults (bottom left panel) (scale 25 μm) and quantification of muscle fibre thickness in larvae following irradiation and in middle aged (18 months) and geriatric zebrafish (>36 months) (right panels). Data shown as mean ± SEM. Each dot represents an animal. (b) Representative example of distance travelled by zebrafish over 30 min in a 24‐well plate at 5 ( |
|
|
|
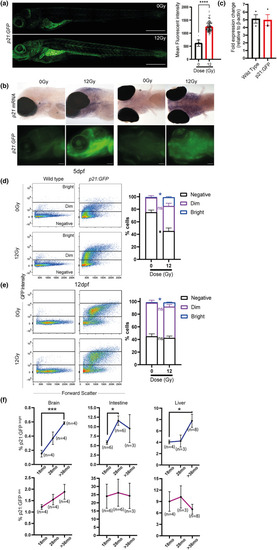
GFPBright cells are associated with other markers of senescence at 5 and 12 dpf. (a) Representative flow cytometry profiles of 5 dpf |
|
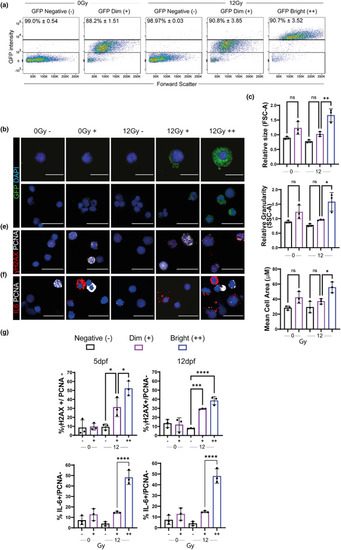
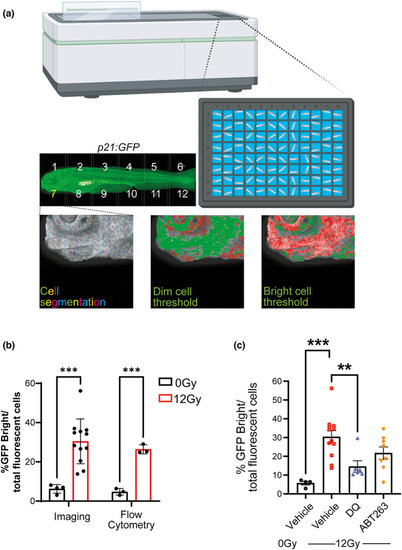
Senolytics reduces the number of p21:GFPBright cells. (a) Diagram representing automated imaging method for p21:GFP zebrafish using The Opera Phenix High‐Content Screening System. Tiled confocal photomicrographs from Opera Phenix microscope of 5 dpf p21:GFP zebrafish were acquired, and individual cells were segregated for analysis. The mean fluorescence intensity of individual cells was classified against a threshold set on the basis of level of fluorescence in wild‐type fish and non‐irradiated p21:GFP fish to identify the GFPBright population; (b) Percentage of GFPBright cells calculated by Opera Phenix High‐Content Imaging and Flow Cytometry analysis, as a proportion of total fluorescent cells in p21:GFP fish with or without irradiation. (c) Quantification of the proportion of GFPBright at 5 dpf in laterally oriented p21:GFP fish following irradiation at 2 dpf and treatment starting at 3 dpf with vehicle, dasatinib (D) plus quercetin (Q) or ABT263 (navitoclax). Data were examined with one‐way ANOVA with Tukey's multiple comparison's test. Data (from two independent experiments are presented as Mean ± SEM presented. *** |