- Title
-
High-throughput functional analysis of autism genes in zebrafish identifies convergence in dopaminergic and neuroimmune pathways
- Authors
- Weinschutz Mendes, H., Neelakantan, U., Liu, Y., Fitzpatrick, S.E., Chen, T., Wu, W., Pruitt, A., Jin, D.S., Jamadagni, P., Carlson, M., Lacadie, C.M., Enriquez, K.D., Li, N., Zhao, D., Ijaz, S., Sakai, C., Szi, C., Rooney, B., Ghosh, M., Nwabudike, I., Gorodezky, A., Chowdhury, S., Zaheer, M., McLaughlin, S., Fernandez, J.M., Wu, J., Eilbott, J.A., Vander Wyk, B., Rihel, J., Papademetris, X., Wang, Z., Hoffman, E.J.
- Source
- Full text @ Cell Rep.
|
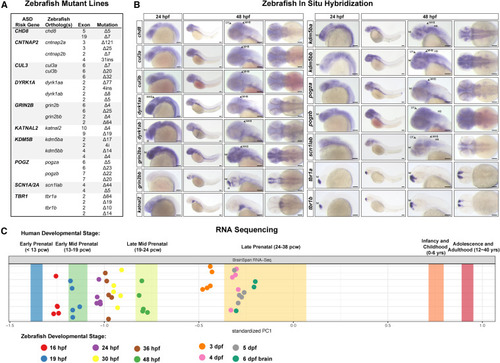
ASD gene expression in zebrafish and mutant generation (A) Zebrafish mutant lines. See Table S and Figures S1A–S1I for mutant sequences. (B) Whole-mount RNA in situ hybridization of ASD gene orthologs in zebrafish at 24 and 48 hpf. For cntnap2a and cntnap2b, see Hoffman et al. (2016).21 Lateral views are shown in the left three panels; dorsal views are shown in the right panel. Anterior to the left; tel, telencephalon; di, diencephalon; MHB, midbrain-hindbrain boundary; OT, optic tectum; HB, hindbrain. Scale bars, 0.1 mm. (C) Mapping zebrafish RNA-seq datasets30 to the human BrainSpan developmental transcriptome RNA-seq dataset 31 using principal component analysis.13 Principal component 1 (PC1) is plotted. Bands represent the 95% confidence interval of the mean PC1 value for human developmental stages. Biological replicates of zebrafish samples are shown as points. pcw, postconception weeks; hpf, hours post-fertilization; dpf, days post-fertilization. See Figure S1L for the full PCA |
|
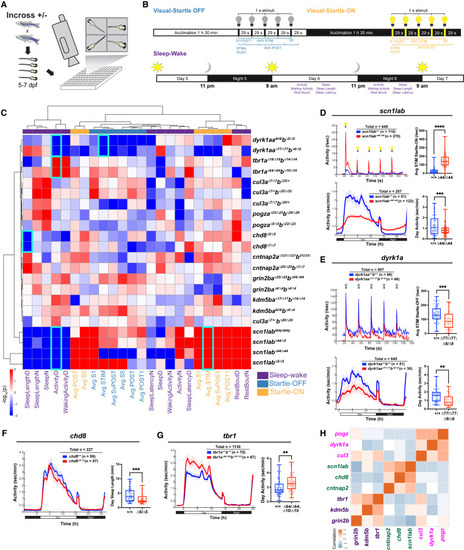
Behavioral phenotypes in zebrafish ASD gene mutants (A and B) Experimental setup and behavioral assays. Visual-startle: Larvae are exposed to five 1-s flashes of lights-off or -on stimuli at 29-s intervals at 5 dpf. Sleep-wake: Larvae are exposed to a 14 h:10 h white light:dark schedule at 5–7 dpf. Visual-startle and sleep-wake parameters are shown (see STAR Methods). (C) Hierarchical clustering of mutant behavioral fingerprints. Each rectangle represents the signed -log10-transformed p values from linear mixed models (LMM) comparing mutant and background-matched wild-type fish (red, increased in mutant; blue, decreased in mutant). Behavioral parameters from startle-off (light blue), startle-on (orange), and sleep-wake (purple) are shown. All mutant lines are shown except katnal2, which lacks consistently significant behavioral features (Figure S2A). Three mutant lines andtransheterozygotes (scn1labΔ44/Δ5) are shown for scn1lab. Behavioral features shown in individual graphs (D–G) are outlined in cyan. Mean and SD values for all groups and beta and p values for all behavioral parameters are shown in Table S2. (D–G) Significant behavioral phenotypes in ASD gene mutants: increased responses to lights-on stimuli, daytime hypoactivity, and night-time hyperactivity in scn1labΔ44/Δ44 (D); decreased responses to lights-off stimuli and daytime hypoactivity in dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (E); decreased daytime sleep bout length in chd8Δ5/Δ5 (F); and daytime hyperactivity in tbr1aΔ64/Δ64tbr1bΔ10/Δ10 (G). Sleep-wake time series graphs in (D–G) show a rolling average of activity data points every 50 min. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01 (one-way ANOVA). (H) Correlation analysis across all 24 behavioral parameters identifies three sub-groups with related phenotypes: (i) scn1labΔ44/Δ44, chd8Δ5/Δ5, cntnap2aΔ121/Δ121cntnap2b31ins/31ins; (ii) pogzaΔ23/Δ23pogzbΔ20/Δ20, dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8, cul3aΔ7/+cul3bΔ32/Δ32; and (iii) tbr1aΔ64/Δ64tbr1bΔ10/Δ10, kdm5baΔ17/Δ17kdm5bbΔ14/Δ14, grin2baΔ25/Δ25grin2bbΔ64/Δ64. |
|
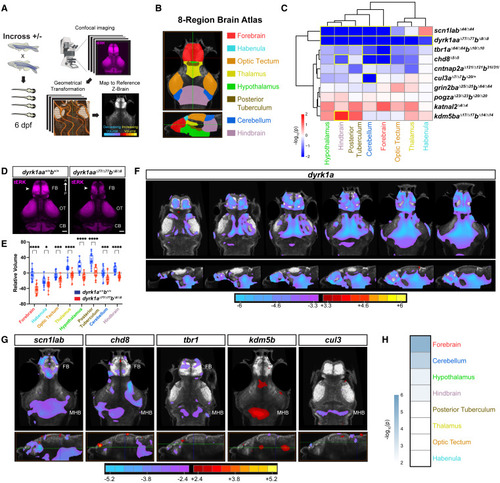
Zebrafish mutants of ASD genes display brain size phenotypes (A) Whole-brain volume mapping pipeline (STAR Methods). (B) Eight-region zebrafish brain atlas derived from Thyme et al. (2019) and Randlett et al. (2015).22,42 (C) Hierarchical clustering of regional brain volume measurements comparing mutant and background-matched wild-type fish. Each rectangle in the clustergram represents the signed -log10-transformed p values from linear mixed models (LMM) (red, increased in mutant; blue, decreased in mutant). Regions with significant volume differences by LMM (p < 0.05) are outlined in yellow. p-values for homozygous and heterozygous mutants are shown in Figure S3A. For raw volume quantifications by region for all fish, see Table S3. (D) tERK-immunostained brains of dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 and wild-type larvae at 6 dpf. Note the decrease in forebrain volume in mutants (arrowheads). Dorsal views. FB, forebrain; OT, optic tectum; CB, cerebellum. Scale bar, 50 μm. (E) Regional brain volume differences in dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (n = 18) and wild-type (n = 18) relative to the standard zebrafish reference brain 42 (dotted line). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗p < 0.05 (one-way ANOVA). (F) Voxel-wise Z score values representing brain volume differences in dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 versus background-matched wild-type larvae. Images shown from left to right represent sequential slices: axial views, top row, dorsal to ventral; sagittal views, bottom row, lateral to medial. Scale bar, Z score (red/yellow, increased in mutant; cyan/purple, decreased in mutant). (G) Voxel-wise Z score values representing brain volume differences for the following lines: scn1labΔ44/Δ44, chd8Δ5/Δ5, tbr1aΔ64/Δ64tbr1bΔ10/Δ10, kdm5baΔ17/Δ17kdm5bbΔ14/Δ14, and cul3aΔ7/Δ7cul3bΔ20/+. Axial views, top row; sagittal views, bottom row. The horizontal green line in the sagittal view indicates the slice shown in the axial view. FB, forebrain; MHB, midbrain-hindbrain boundary. Scale bar, Z score (red/yellow, increased in mutant; cyan/purple, decreased in mutant). For the number of animals used in imaging experiments, see STAR Methods (average n = 23–26 per genotype in two to four independent clutches). (H) Genotype-level p values for each brain region combined for all homozygous mutants and cul3aΔ7/Δ7cul3bΔ20/+, excluding dyrk1a, using Fisher’s method. Scale represents the -log10-transformed combined p value. The forebrain (telencephalon) shows the most significant difference in volume (p = 8.32 × 10−6, Fisher’s combined probability test). |
|
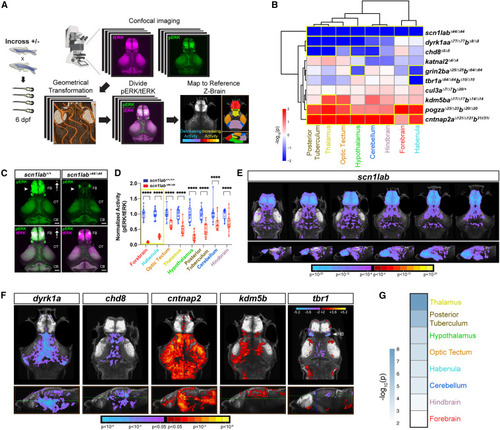
Altered baseline brain activity in zebrafish ASD gene mutants (A) Whole-brain activity mapping pipeline (STAR Methods). (B) Hierarchical clustering of regional brain activity (pERK/tERK) in mutant versus background-matched wild-type fish using linear mixed models (LMM). Each rectangle in the clustergram represents the signed -log10-transformed p values from LMM (red, increased in mutant; blue, decreased in mutant). Regions with significant activity differences by LMM (p < 0.05) are outlined in yellow. p-values for all homozygous and heterozygous mutants are shown in Figure S4A. For pERK/tERK ratios by region for all fish, see Table S4. (C) pERK- (top) and merged pERK- and tERK-immunostained brains of scn1labΔ44/Δ44 and wild-type larvae at 6 dpf. Note the decrease in pERK staining in the mutant forebrain (arrowheads). Dorsal views. FB, forebrain; OT, optic tectum; CB, cerebellum. Scale bar, 50 μm. (D) Normalized regional activity (pERK/tERK) in scn1labΔ44/Δ44 (n = 23) and background-matched wild-type (n = 21). ∗∗∗∗p < 0.0001 (one-way ANOVA). (E) Voxel-wise Z score pERK/tERK values representing brain activity differences in scn1labΔ44/Δ44 versus background-matched wild-type larvae. Images shown from left to right represent sequential slices: axial views, top row, dorsal to ventral; sagittal views, bottom row, lateral to medial. Scale bar, Z score (red/yellow, increased in mutant; cyan/purple, decreased in mutant). Results are shown at p < 0.05 whole-brain family-wise error (FWE) corrected with an initial p threshold of 0.0001. (F) Voxel-wise Z score pERK/tERK values representing brain activity differences for the following lines: dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8, chd8Δ5/Δ5, cntnap2aΔ121/Δ121cntnap2b31ins/31ins, kdm5baΔ17/Δ17kdm5bbΔ14/Δ14, and tbr1aΔ64/Δ64tbr1bΔ10/Δ10. Axial views, top row; sagittal views, bottom row. The horizontal green line in the sagittal view indicates the slice shown in the axial view. Scale bar represents Z score (red/yellow, increased in mutant; cyan/purple, decreased in mutant). Results are shown at p < 0.05 whole-brain FWE corrected with an initial p threshold of 0.05 for all mutants except tbr1, which is not FWE corrected because significance did not meet cluster correction. tbr1 mutants display significantly decreased activity in the habenula (HB) (arrowhead) by LMM (B). For the number of animals used in imaging experiments, see STAR Methods (average n = 22–24 per genotype in two to three independent clutches). (G) Genotype-level p values for each brain region are combined for all homozygous mutants and cul3aΔ7/Δ7cul3bΔ20/+, excluding scn1lab, using Fisher’s method. The scale represents the -log10-transformed combined p value. The thalamus shows the most significant difference in activity (p = 9.55 × 10−9, Fisher’s combined probability test). |
|
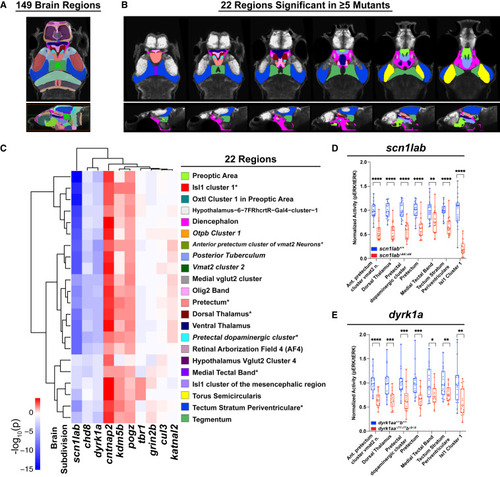
Mutant activity phenotypes converge on 22 brain regions (A) The 149-region zebrafish brain atlas derived from Randlett et al. (2015).42 (B) The 22 regions with significant differences in baseline brain activity in at least five mutants. Region names are shown in (C). Images shown from left to right represent sequential slices: axial views, top row, dorsal to ventral; sagittal views, bottom row, lateral to medial. (C) Hierarchical clustering of 22 regions with significant differences in baseline brain activity in at least five mutants by linear mixed models (LMM) (p < 0.05). Five regions involve dopaminergic signaling (italics): posterior tuberculum, pre-tectal dopaminergic cluster, vmat2 cluster 2, anterior pretectum cluster of vmat2 neurons, otpb cluster 1. Colors shown to next to each region name refer to (B). ∗The seven regions with significant differences in baseline activity in at least six mutants (Figures S5B and S5C). Hierarchical clustering of 131 regions with significant differences in activity in at least two mutants by LMM is shown in Figure S5A. For a list of significant regions by mutant, see Table S5. (D and E) Normalized brain activity (pERK/tERK) in scn1labΔ44/Δ44 (D) and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (E) versus background-matched wild-type fish in the seven brain regions that show significant differences in at least six mutants. Heterozygous phenotypes are shown in Figures S5D and S5E. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05 (one-way ANOVA). |
|
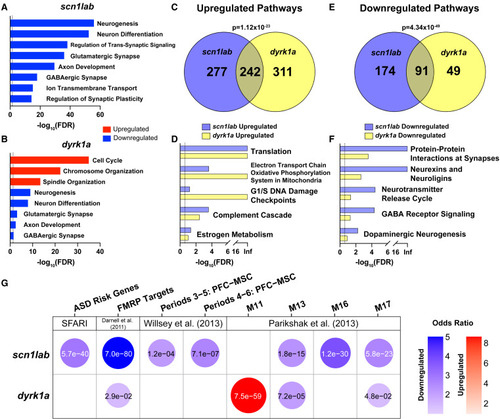
Whole-brain RNA-seq identifies common dysregulated pathways in scn1lab and dyrk1a mutants (A and B) Dysregulated GO pathways in scn1labΔ44/Δ44 (A) and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (B) mutants. The dashed line represents FDR of less than 0.05. Pathways enriched in upregulated (red) or downregulated (blue) genes are shown. For a complete list of significant GO pathways, see Table S6. (C–F) GSEA pathways upregulated (C and D) or downregulated (E and F) in both mutants. The dashed line represents FDR of less than 0.25. p values indicating significant overlap were calculated using Fisher’s exact test. For a complete list of significant GSEA pathways, see Table S6. (G) Hypothesis-driven GSEA of DE genes (p < 0.1 and fold-change >1.5) in scn1labΔ44/Δ44 and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 mutants using the following datasets: SFARI ASD risk genes,32 FMRP targets,51 and human brain co-expression network modules.8,9 The top four modules from Parikshak et al. (2013) 9 based on combined p value in homozygous mutants using Fisher’s method are shown. Gene sets enriched in upregulated (red) or downregulated (blue) genes are shown. Bubbles are shown only for gene sets with significant enrichment (p < 0.05). The color intensity and size of each bubble represent the odds ratio. p-values calculated using Fisher’s exact test are shown in each bubble. For the complete GSEA including all co-expression network modules in homozygotes and heterozygotes, see Figures S6B and S6C. |
|
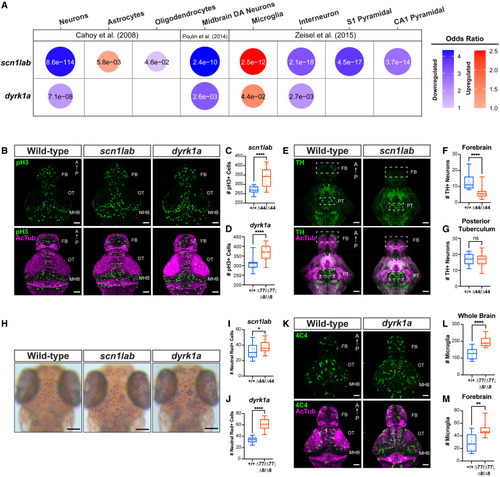
Dopaminergic and microglial phenotypes in scn1lab and dyrk1a mutants (A) Hypothesis-driven GSEA of DE genes (p < 0.1 and fold-change >1.5) in scn1labΔ44/Δ44 and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 mutants using cell-type-specific datasets.58,59,60 Cell type markers enriched in upregulated (red) or downregulated (blue) genes are shown. Bubbles are shown only for cell type markers with significant enrichment (p < 0.05). The color intensity and size of each bubble represent the odds ratio. p-values calculated using Fisher’s exact test are shown in each bubble. For the complete GSEA in homozygotes and heterozygotes, see Figure S7A. (B) Phospho-histone H3 (pH3, green) and acetylated tubulin (AcTub, magenta) immunostained whole brains of scn1labΔ44/Δ44 and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 versus wild-type fish at 3 dpf. Dorsal views. FB, forebrain; OT, optic tectum; MHB, midbrain-hindbrain boundary. Scale bar, 50 μm. For quantification by brain region, see Figures S7B–S7E. (C and D) Total number of pH3+ cells in scn1labΔ44/Δ44 (n = 18) versus scn1lab+/+ (n = 16) (C) and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (n = 14) versus wild-type fish (n = 19) (D). ∗∗∗∗p < 0.0001, one-way ANOVA. (E) Tyrosine hydroxylase (TH, green) and acetylated tubulin (AcTub, magenta) immunostained whole brains of wild-type, scn1labΔ44/Δ44, and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 fish at 4 dpf. Scale bar, 50 μm. Ventral views. FB, forebrain; PT, posterior tuberculum. (F and G) Total number of TH+ cells in scn1labΔ44/Δ44 (n = 20) versus scn1lab+/+ (n = 19) in the forebrain (F) and posterior tuberculum (G) (boxes in [E]). ∗∗∗∗p < 0.0001, one-way ANOVA. For heterozygous phenotypes, see Figures S7F–S7G. (H) Neutral red staining of live scn1labΔ44/Δ44 and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 versus wild-type fish at 4 dpf. Scale bar, 0.1 mm. (I and J) Total number of neutral red+ cells in scn1labΔ44/Δ44 (n = 17) versus scn1lab+/+ (n = 20) (I) and dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (n = 16) versus wild-type fish (n = 15) (J). ∗∗∗∗p < 0.0001, ∗p < 0.05, one-way ANOVA. (K) 4C4 (green) and acetylated tubulin (AcTub, magenta) immunostaining of whole brains of dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 and wild-type fish at 4 dpf. Scale bar, 50 μm. Dorsal views. FB, forebrain; OT, optic tectum; MHB, midbrain-hindbrain boundary. (L and M) Total number of 4C4+ cells in dyrk1aaΔ77/Δ77dyrk1abΔ8/Δ8 (n = 13) versus wild-type (n = 10) in whole brain (L) and forebrain (M). ∗∗∗∗p < 0.0001, ∗∗p < 0.01, one-way ANOVA. |