- Title
-
Dissecting cell identity via network inference and in silico gene perturbation
- Authors
- Kamimoto, K., Stringa, B., Hoffmann, C.M., Jindal, K., Solnica-Krezel, L., Morris, S.A.
- Source
- Full text @ Nature
|
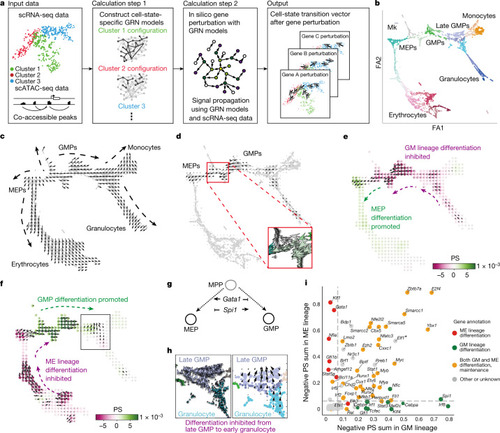
Overview of CellOracle and application to haematopoiesis.
a, Simulation of cell-state transitions in response to TF perturbation. First, CellOracle constructs custom transcriptional GRNs using scRNA-seq and scATAC-seq data (left). Accessible promoter and enhancer peaks from scATAC-seq data are then combined with scRNA-seq data to generate cluster-specific GRN models (middle). CellOracle simulates the change in cell state in response to a TF perturbation, projecting the results onto the cell trajectory map (right). b, Force-directed graph of 2,730 myeloid progenitor cells from Paul et al.16. Twenty-four cell clusters (Louvain clustering) were organized into six main cell types. Mk, megakaryocytes. c, Differentiation vectors for each cell projected onto the force-directed graph. d, CellOracle simulation of cell-state transition in Spi1 KO simulation. Summarized cell-state transition vectors projected onto the force-directed graph. Vectors for each cell are shown in the inset. e, Spi1 KO simulation vector field with perturbation scores (PSs). f, Gata1 KO simulation with perturbation scores. g, Schematic of Spi1–Gata1 lineage switching. MPP, multipotent progenitor. h, Detail of Gata1 simulation for the granulocyte branch. Left, cell-state transition vectors for each cell. Right, summarized vectors. i, Systematic KO simulation result of 90 TFs in the GM and ME lineage is summarized as a scatter plot of the sum of negative perturbation scores (shown in log scale). Dashed lines represent cut-off values corresponding to false-positive rate (FPR) = 0.01. Genes are classified into four categories on the basis of their previously reported functions (Supplementary Table 2). The asterisk refers to Supplementary Fig. 11, where we expand on the predicted phenotype. All scores can be explored through our web application (https://celloracle.org). |
|
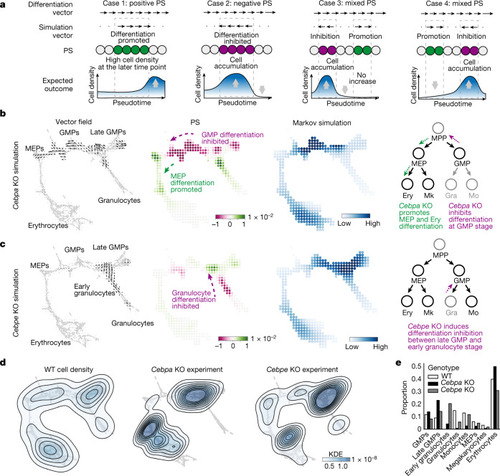
Validation of CellOracle using experimentally measured cell density in Cebpa and Cebpe KOs in haematopoiesis.
a, Biological interpretation of perturbation scores (estimation of cell density based on perturbation score). Case 1: the differentiation and perturbation simulation vectors share the same direction, indicating a population shift towards a more differentiated identity. Case 2: the two vectors are opposed, suggesting that differentiation is inhibited. Case 3: predicted inhibition precedes promotion; thus, cells will be likely to accumulate. b,c, CellOracle Cebpa KO (b) and Cebpe KO (c) simulations showing cell-state transition vectors, perturbation scores and estimated cell density (Markov simulation). Right, schematics of simulated phenotype. Ery, erythrocyte. d, Ground-truth experimental cell density plot of wild-type (WT) cells, Cebpa KO cells and Cebpe KO cells in the force-directed graph embedding space. Estimated kernel density data are shown as a contour line on a scatter plot to depict cell density. e, Cell-type proportions in the WT and ground-truth KO samples. Gra, granulocyte; KDE, kernel density estimation; Mo, monocyte. |
|
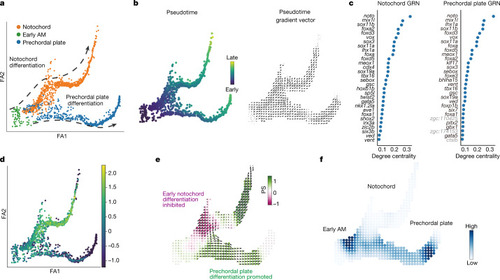
CellOracle KO simulation with zebrafish embryogenesis data.
a, Two-dimensional force-directed graph of the axial mesoderm (AM) sub-branch (n = 1,669 cells) in a published zebrafish embryogenesis atlas (Farrell et al.32). Arrows indicate notochord cell differentiation (top) and prechordal plate differentiation (bottom). b, Conversion of URD-calculated pseudotime (left) into a 2D pseudotime gradient vector field (right). c, Degree centrality scores were used to rank the top 30 TFs in notochord (left) and prechordal plate (right). Black text denotes TFs. Grey text denotes non-TFs. d, Expression of noto projected onto the axial mesoderm sub-branch. e, Noto KO simulation vector and perturbation scores. f, Markov simulation to estimate cell density in the noto KO sample. The simulation predicted inhibited early notochord differentiation and promotion of prechordal plate differentiation, indicating a potential lineage switch. |
|
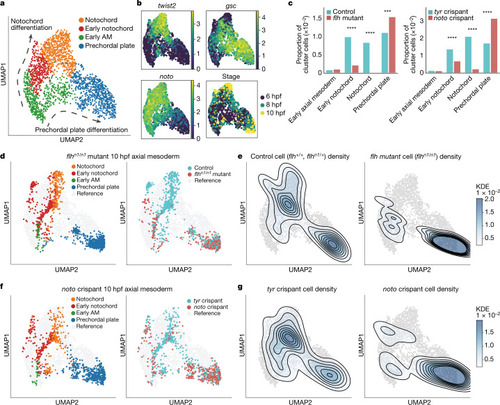
Experimental validation of zebrafish noto LOF predictions.
a, UMAP plot of WT reference data for axial mesoderm (6, 8 and 10 hpf): notochord, early notochord, early axial mesoderm and prechordal plate clusters (n = 2,012 cells). Arrows indicate notochord differentiation (top) and prechordal plate differentiation (bottom). b, Gene expression (log-transformed unique molecular identifier (UMI) count) and developmental stage are projected onto the axial mesoderm UMAP plot. Noto and twist2 are expressed in notochord, whereas gsc marks the prechordal plate. c, Bar plots comparing cell cluster compositions between treatments and controls (left, flhn1/n1 mutants (10 hpf) and controls; right, noto crispants (10 hpf) and tyr crispants). Cluster compositions are presented as the proportion of each group normalized to the whole cell number. In both flhn1/n1 mutants and noto crispants, the notochord is significantly depleted (flhn1/n1: P = 5.55 × 10−52; noto: P = 1.39 × 10−33, chi-square test) and the prechordal plate is significantly expanded (flhn1/n1: P = 1.07 × 10−4; noto: P = 5.01 × 10−18, chi-square test. ***P < 0.001; ****P < 0.0001). d–g, flhn1/n1 mutant or noto crispant data projected onto the WT axial mesoderm UMAP plot. d, Cluster annotation and sample label projected onto the UMAP plot. e, Kernel cell density contour plot shows control cell density (left) and flhn1/n1 mutant cell density (right). f, Cluster annotation and sample label projected onto the UMAP plot. g, tyr crispant cell density (left) and noto crispant cell density (right) shown on the kernel cell density contour plot. |
|
Experimental validation of lhx1a as a putative regulator of zebrafish axial mesoderm development.
a, Top 30 TFs according to predicted KO effects. Red and *: previously reported notochord regulators (Supplementary Table 2). lhx1a, sebox and irx3a were selected for experimental validation. b, lhx1a LOF simulation in the axial mesoderm sub-branch, predicting an inhibition of axial mesoderm differentiation from early stages. c, scRNA-seq validation of experimental LOF: cell cluster composition of the axial mesoderm clusters normalized to the whole cell number in lhx1a and tyr (control) crispant samples. Early notochord is significantly expanded (P = 1.34 x 10−35, chi-square test) and differentiated axial mesoderm populations are significantly depleted (notochord: P = 3.83 x 10−3; prechordal plate: P = 1.28 x 10−7, chi-square test) in lhx1a crispants. d, lhx1a and tyr crispant axial mesoderm cells at 10 hpf. Left, cell type annotation of lhx1 and tyr crispant cells. Right, lhx1a and control crispant data projected onto the WT UMAP. e, Control cell density (left, n = 2,342 cells) and lhx1a crispant cell density (right, n = 2,502 cells). f, Rug plot showing the difference in averaged NMF module scores between lhx1a and tyr crispants in notochord lineage cells. Black, cell-type-specific modules. Light grey, broad cluster modules. CM, cephalic mesoderm. g, Violin plot of NMF module score in notochord lineage cells (n = 1,918 lhx1a crispant and n = 2,616 tyr crispant cells. h, Violin plots of gene expression in the notochord (NC) lineage cells. ****P < 0.0001, two-tailed Wilcoxon rank-sum test with Bonferroni correction. i, Quantification (number of spots in flattened HCR image) normalized to WT. Mean ± s.e.m. n = 2 independent biological replicates, 8 embryos per replicate. nog1: P = 0.0022 (WT versus lhx1a crispant), P = 0.0052 (tyr versus lhx1a crispant); gsc: P = 0.00042 (WT versus lhx1a crispant), P = 0.0018 (tyr versus lhx1a crispant); twist2: P = 0.0011 (WT versus lhx1a crispant), P = 0.0012 (tyr versus lhx1a crispant); two-sided t-test. j, Representative HCR images for nog1 expression (yellow) in whole embryos at 10 hpf. k, Representative flattened HCR images of 10 hpf embryos stained with probes against gsc (yellow) and twist2 (red); nuclei are stained with DAPI (blue). Scale bars, 300 μm. |