- Title
-
Single-cell transcriptomes and runx2b-/- mutants reveal the genetic signatures of intermuscular bone formation in zebrafish
- Authors
- Nie, C.H., Wan, S.M., Chen, Y.L., Huysseune, A., Wu, Y.M., Zhou, J.J., Hilsdorf, A.W.S., Wang, W.M., Witten, P.E., Lin, Q., Gao, Z.X.
- Source
- Full text @ Natl Sci Rev
|
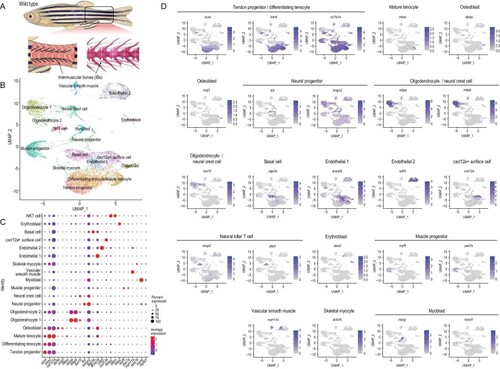
Global patterns of single-cell expression profiles of the tissue of origin of intermuscular bones (IBs) in wild-type zebrafish, and identification of cell types. (A) Pattern of tissue collection for scRNA-seq. (B) Uniform manifold approximation and projection (UMAP) visualization of 18 subsets from 13 |
|
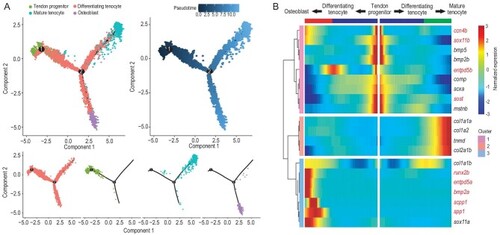
Differentiation trajectory construction of tendons and intermuscular bones, and screening of key regulators for IB formation. (A) Diffusion map visualization of the tendon and IB trajectories simulated by monocle2 across tendon progenitors, mature tenocytes, differentiating tenocytes and osteoblasts on the left. The corresponding diffusion pseudo-time is indicated in the bottom-left frame. A diffusion map visualization of each cluster is shown on the right. (B) Heatmap of the gene expression (smoothed over three adjacent cells) in subsets ordered by the pseudo-time of mature tenocytes and osteoblasts, as in (A); some curated genes are shown. |
|
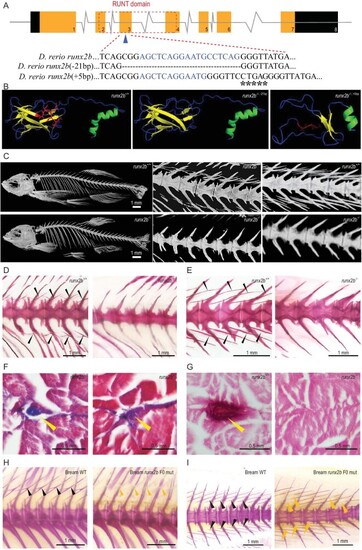
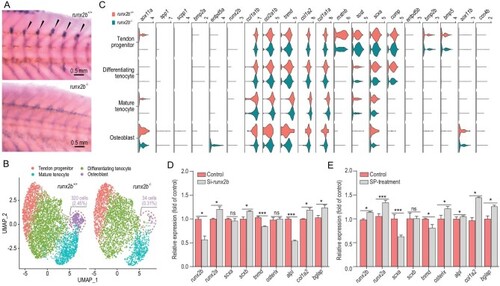
Gene structure and phenotypic characteristics of |
|
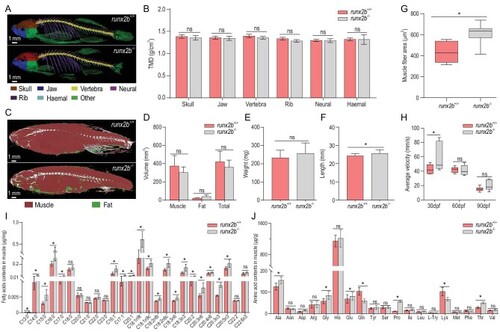
Comparison of characteristics of other bones, swimming performance and muscle nutrient content between |
|
Comparative transcriptome analysis of |