- Title
-
Eomes function is conserved between zebrafish and mouse and controls left-right organiser progenitor gene expression via interlocking feedforward loops
- Authors
- Talbot, C.D., Walsh, M.D., Cutty, S.J., Elsayed, R., Vlachaki, E., Bruce, A.E.E., Wardle, F.C., Nelson, A.C.
- Source
- Full text @ Front Cell Dev Biol
|
Mouse |
|
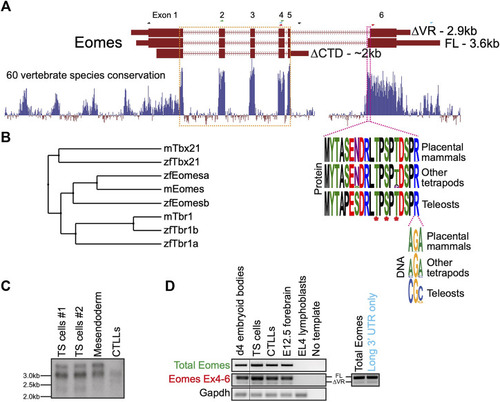
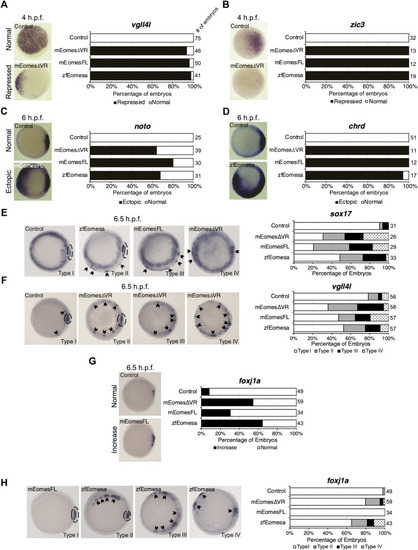
Both FL and ∆VR isoforms of mouse Eomes are functionally equivalent to zebrafish Eomesa in the early embryo. WISH analysis of ectoderm markers |
|
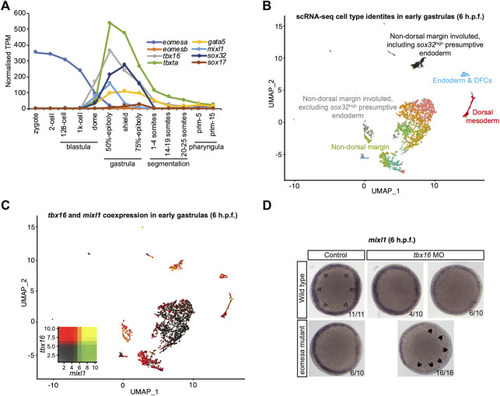
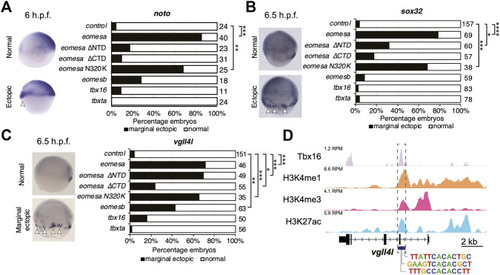
Eomesa and Tbx16 are redundantly required for |
|
Tbx16 is substantially co-expressed with |
|
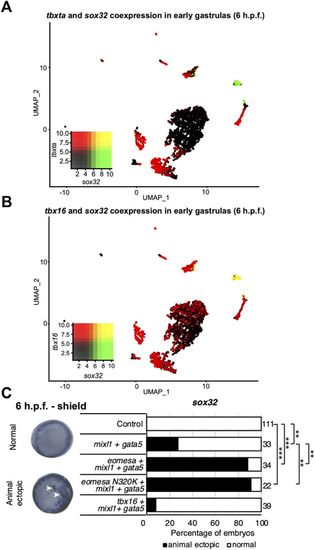
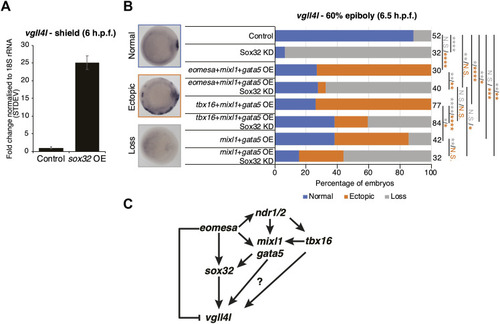
Eomesa is a more potent inducer of endoderm, organiser and dorsal forerunner cell markers than other T-box factors. |
|
Eomesa activation of |