FIGURE 2
- ID
- ZDB-FIG-220923-58
- Publication
- Talbot et al., 2022 - Eomes function is conserved between zebrafish and mouse and controls left-right organiser progenitor gene expression via interlocking feedforward loops
- Other Figures
- All Figure Page
- Back to All Figure Page
|
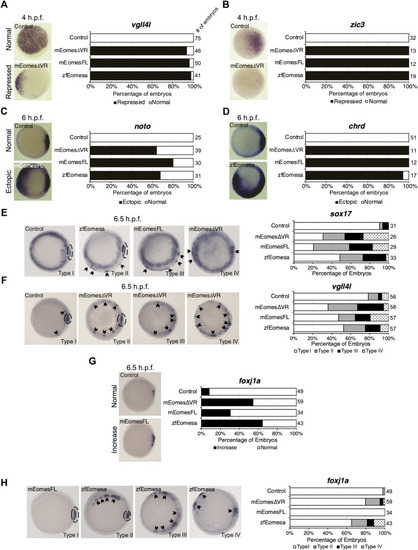
Both FL and ∆VR isoforms of mouse Eomes are functionally equivalent to zebrafish Eomesa in the early embryo. WISH analysis of ectoderm markers |