- Title
-
Endothelial versus pronephron fate decision is modulated by the transcription factors Cloche/Npas4l, Tal1, and Lmo2
- Authors
- Mattonet, K., Riemslagh, F.W., Guenther, S., Prummel, K.D., Kesavan, G., Hans, S., Ebersberger, I., Brand, M., Burger, A., Reischauer, S., Mosimann, C., Stainier, D.Y.R.
- Source
- Full text @ Sci Adv
|
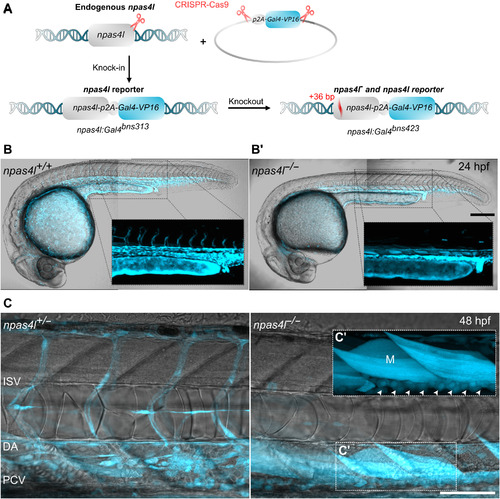
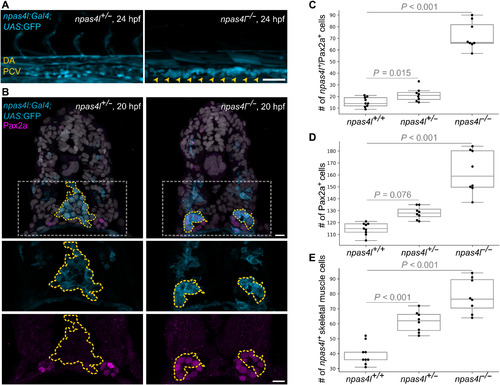
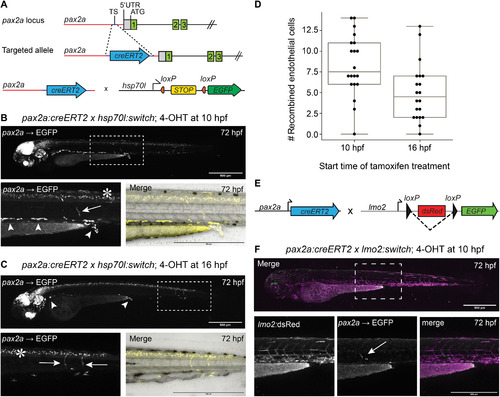
EXPRESSION / LABELING:
PHENOTYPE:
|
|
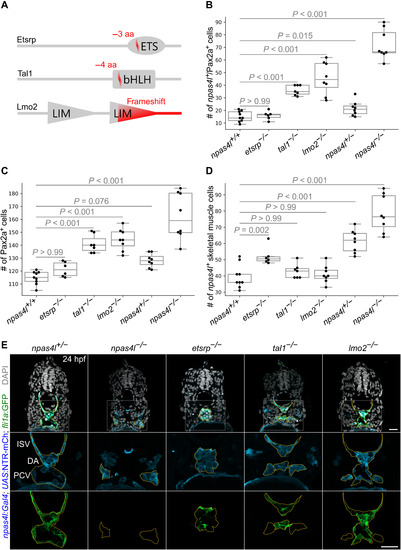
EXPRESSION / LABELING:
PHENOTYPE:
|
|
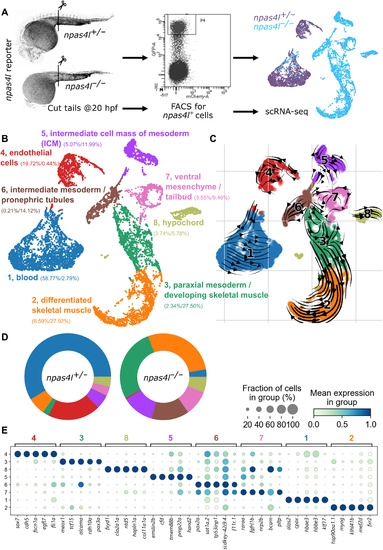
Fig. 3. Transcriptomic profiling of npas4l reporter–expressing cells in npas4l+/− and npas4l−/− embryos. (A) Schematic of the experimental protocol. 5207 and 5262 npas4l reporter–expressing cells from the posterior half of 20 hpf npas4l+/− and npas4l−/− embryos, respectively, were sequenced using the 10x Genomics sequencing platform. FACS, fluorescence-activated cell sorting. (B) Uniform manifold approximation and projection (UMAP) representation of the data clustered by the Leiden algorithm. Numbers between parentheses indicate the percentage of heterozygous/mutant cells contributing to the cluster. (C) Velocity analysis done by comparing pre-mRNA and mRNA levels to infer the relation between the cells in a cluster. (D) Cluster composition per genotype. Graphical representation of the numbers displayed in (B). The main clusters increasing in npas4l−/− compared with npas4l+/− siblings are paraxial mesoderm (cluster 3), skeletal muscle (cluster 2), and intermediate mesoderm (cluster 6). (E) Dot plot with the top five markers separating the clusters. Pseudobulk expression data and marker genes used to separate the clusters can be found in tables S1 and S2, respectively. Differential gene expression in pseudo-bulk analysis can be found in table S3. The full and annotated dataset can be explored at https://bioinformatics.mpi-bn.mpg.de/20hpf_npas4l_expressing_cell or downloaded as an annotated data file using the link https://figshare.com/s/e6bdd5be14c7085d606c. |
|
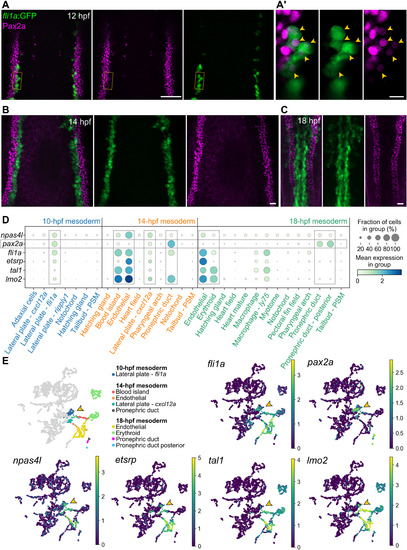
Fig. 4. fli1a and pax2a/Pax2a expression overlap during early stages of wild-type development and largely segregate before 14 hpf. Confocal imaging reveals overlapping expression of fli1a:GFP and Pax2a (A), which decreases by 14 hpf (B), and is not detected at 18 hpf (C). Images of individual 0.74 μm optical sections (A′) clearly suggest colocalization. Yellow arrowheads point to double-positive cells. (D) Mapping of 10 to 18 hpf mesodermal cells from a single-cell time course dataset (39) suggests coexpression of endothelial markers and pax2a at 10 hpf, which decreases over time. PSM, presomitic mesoderm. (E) UMAP plots showing the expression of npas4l, pax2a, fli1a, tal1, lmo2, and etsrp; arrowheads point to the 10 hpf fli1a/pax2a double-positive cluster. Relevant clusters have been color-coded in the left plot in (E) and boxed with dotted lines in (D). “Pronephric duct” = pronephron tubule. Scale bars, 50 μm (A), 10 μm (A′), and 20 μm (B and C). EXPRESSION / LABELING:
PHENOTYPE:
|
|
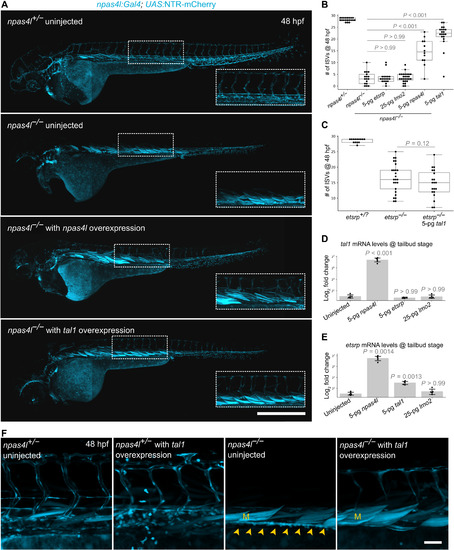
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
|