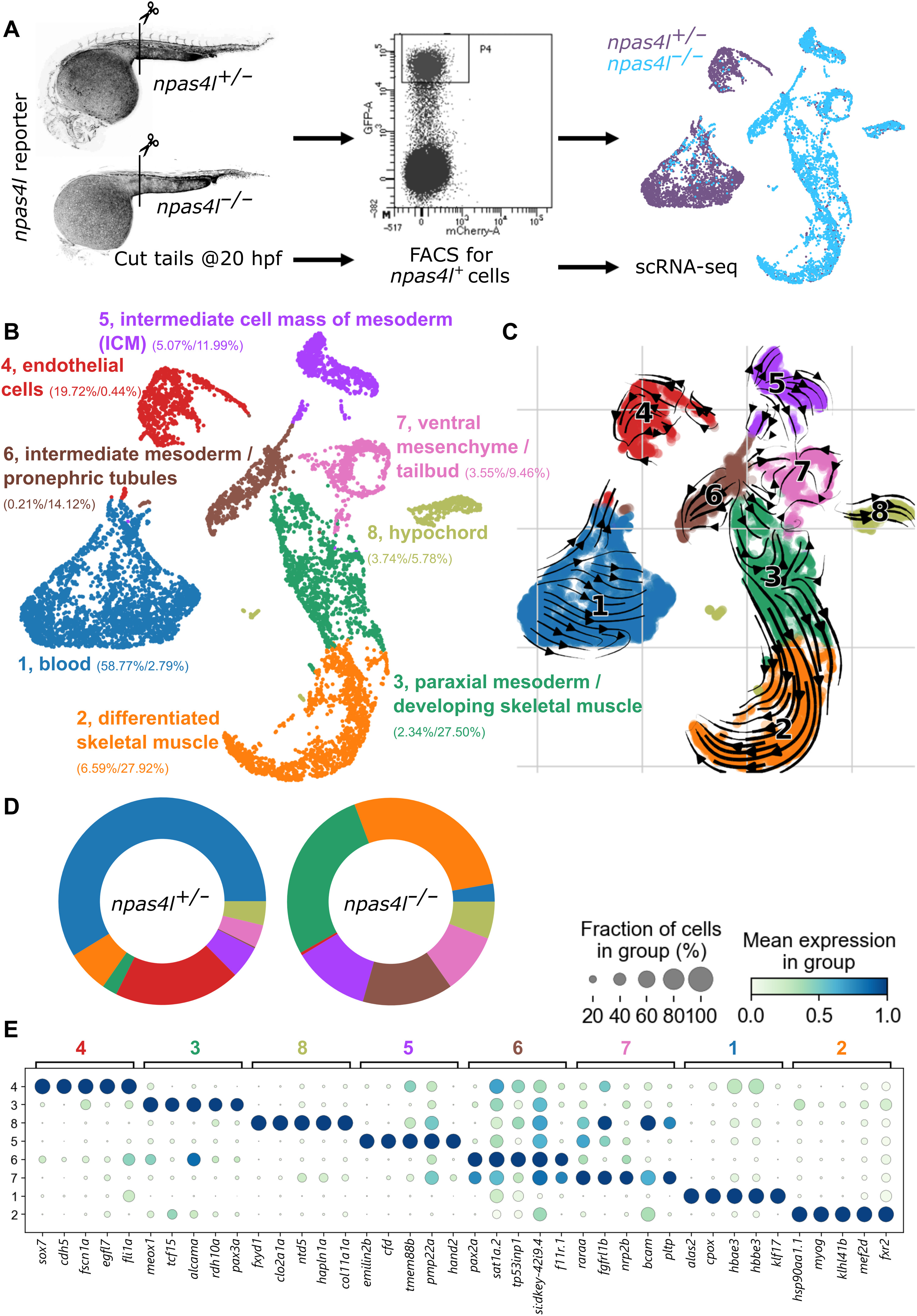
Fig. 3 Fig. 3. Transcriptomic profiling of npas4l reporter–expressing cells in npas4l+/− and npas4l−/− embryos. (A) Schematic of the experimental protocol. 5207 and 5262 npas4l reporter–expressing cells from the posterior half of 20 hpf npas4l+/− and npas4l−/− embryos, respectively, were sequenced using the 10x Genomics sequencing platform. FACS, fluorescence-activated cell sorting. (B) Uniform manifold approximation and projection (UMAP) representation of the data clustered by the Leiden algorithm. Numbers between parentheses indicate the percentage of heterozygous/mutant cells contributing to the cluster. (C) Velocity analysis done by comparing pre-mRNA and mRNA levels to infer the relation between the cells in a cluster. (D) Cluster composition per genotype. Graphical representation of the numbers displayed in (B). The main clusters increasing in npas4l−/− compared with npas4l+/− siblings are paraxial mesoderm (cluster 3), skeletal muscle (cluster 2), and intermediate mesoderm (cluster 6). (E) Dot plot with the top five markers separating the clusters. Pseudobulk expression data and marker genes used to separate the clusters can be found in tables S1 and S2, respectively. Differential gene expression in pseudo-bulk analysis can be found in table S3. The full and annotated dataset can be explored at https://bioinformatics.mpi-bn.mpg.de/20hpf_npas4l_expressing_cell or downloaded as an annotated data file using the link https://figshare.com/s/e6bdd5be14c7085d606c.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci Adv