- Title
-
Stage Specific Transcriptomic Analysis and Database for Zebrafish Oogenesis
- Authors
- Bogoch, Y., Jamieson-Lucy, A., Vejnar, C.E., Levy, K., Giraldez, A.J., Mullins, M.C., Elkouby, Y.M.
- Source
- Full text @ Front Cell Dev Biol
|
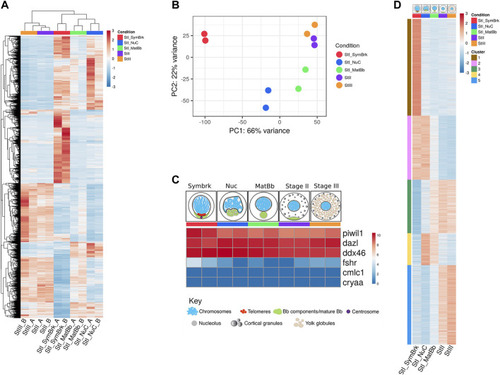
A high throughput analysis of 5 stages of oocyte development. |
|
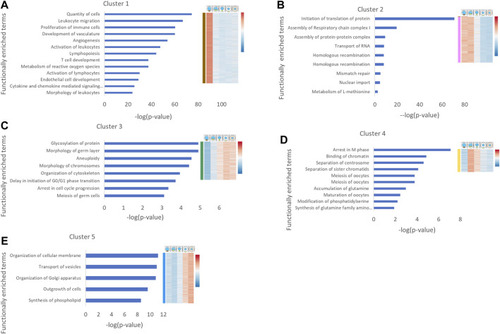
Functional enrichment analysis of gene expression clusters in oogenesis. Human homologues of genes from each cluster were analyzed by IPA. Cancer related functions were removed. The main functionally enriched terms for each cluster are shown. The |
|
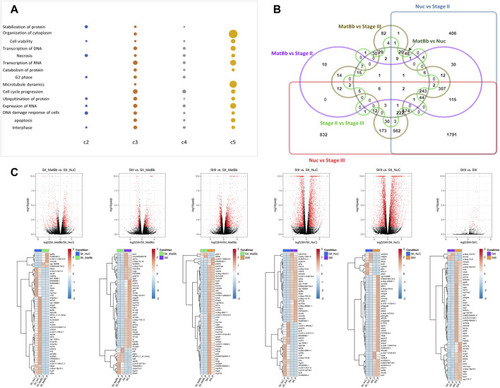
Analysis of differentially expressed genes of different stages. |
|
Functional analysis of pairwise comparisons. |