- Title
-
Transcriptomic underpinnings of high and low mirror aggression zebrafish behaviours
- Authors
- Reichmann, F., Pilic, J., Trajanoski, S., Norton, W.H.J.
- Source
- Full text @ BMC Biol.
|
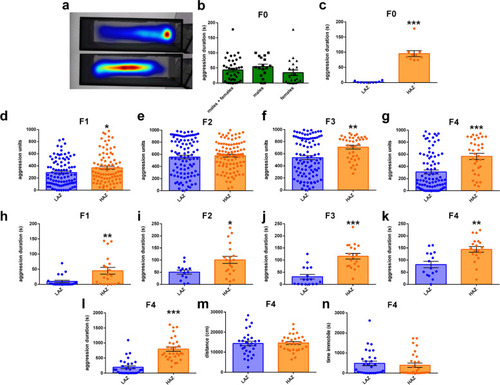
Transgenerational effects of selective breeding on mirror fighting. |
|
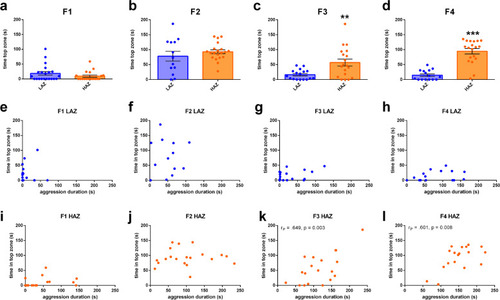
Transgenerational effects of selective breeding on anxiety-like behaviour. |
|
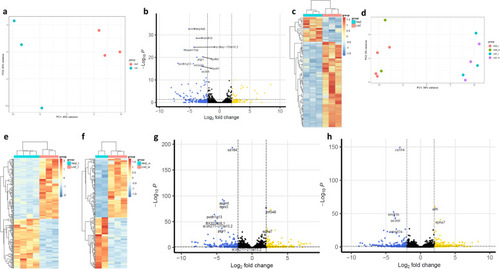
Selective breeding-induced and mirror-induced neurotranscriptomic differences of male and female high aggression zebrafish (HAZ) and low aggression zebrafish (LAZ). |
|
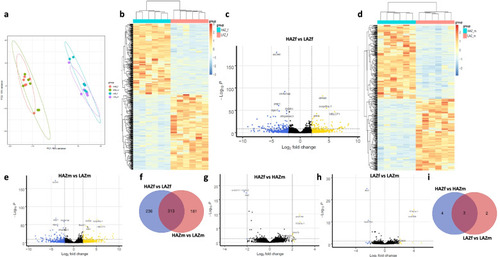
Neurotranscriptomic differences of the most aggressive male and female high aggression zebrafish (HAZ) compared to the least aggressive low aggression zebrafish (LAZ). |
|
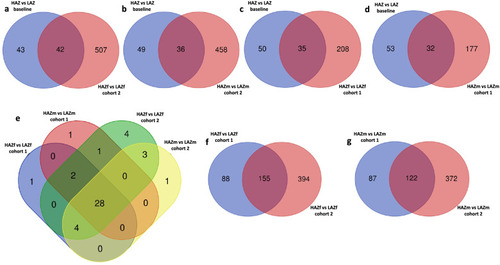
Overlap of differentially expressed genes (DEGs) between high aggression zebrafish (HAZ) and low aggression zebrafish (LAZ) at baseline and after mirror exposure. |
|
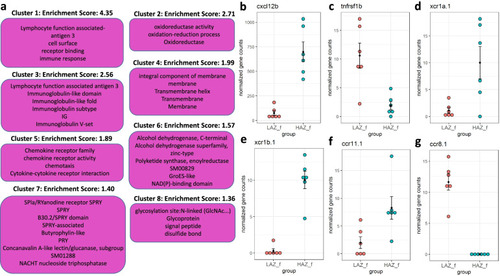
Enriched clusters and functional categories between the most aggressive female high aggression zebrafish (HAZf) and the least aggressive female low aggression zebrafish (LAZf). |
|
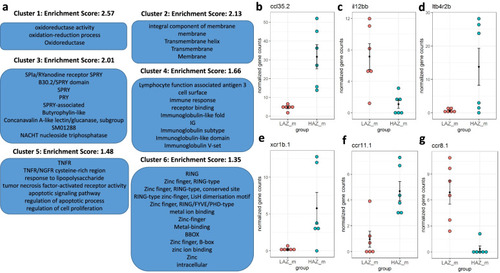
Enriched clusters and functional categories between the most aggressive male high aggression zebrafish (HAZm) and the least aggressive male low aggression zebrafish (LAZm). |
|
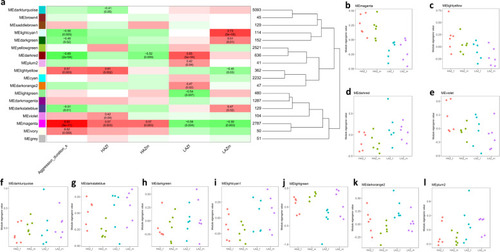
Weighted Gene Coexpression Network Analysis (WGCNA) of the most aggressive high aggression zebrafish (HAZ) compared to the least aggressive low aggression zebrafish (LAZ). |
|
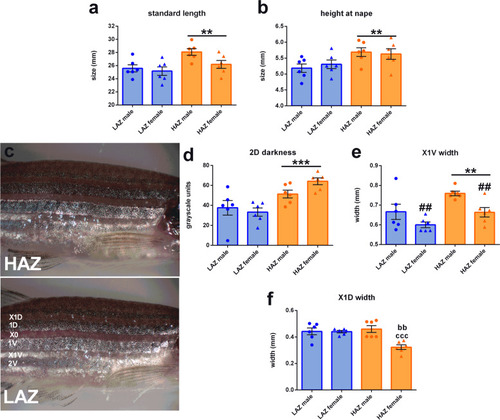
Morphological differences between high aggression zebrafish (HAZ) and low aggression zebrafish (LAZ). |