- Title
-
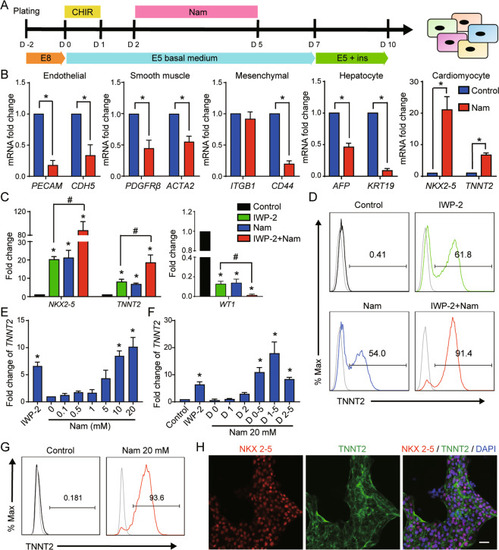
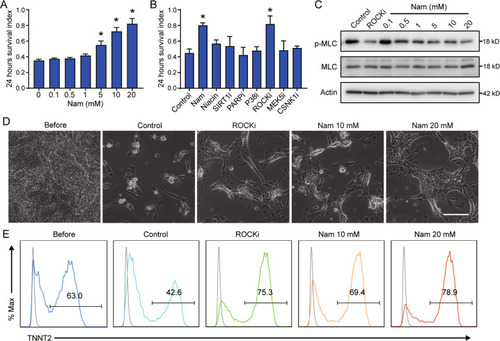
Nicotinamide promotes cardiomyocyte derivation and survival through kinase inhibition in human pluripotent stem cells
- Authors
- Meng, Y., Song, C., Ren, Z., Li, X., Yang, X., Ai, N., Yang, Y., Wang, D., Zhan, M., Wang, J., Lei, C.L., Liu, W., Ge, W., Lu, L., Chen, G.
- Source
- Full text @ Cell Death Dis.
|
|
|
|
|
|
|
|
|
|
|
|
|
PHENOTYPE:
|