- Title
-
Brain Regeneration Resembles Brain Cancer at Its Early Wound Healing Stage and Diverges From Cancer Later at Its Proliferation and Differentiation Stages
- Authors
- Demirci, Y., Heger, G., Katkat, E., Papatheodorou, I., Brazma, A., Ozhan, G.
- Source
- Full text @ Front Cell Dev Biol
|
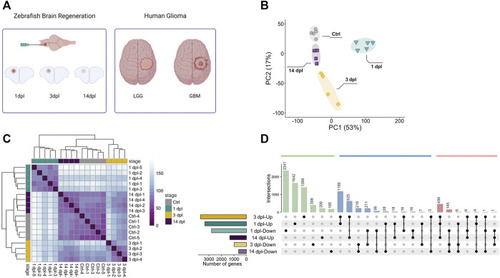
Sample preparation from three different stages of zebrafish brain regeneration and initial analyses of the transcriptome data. |
|
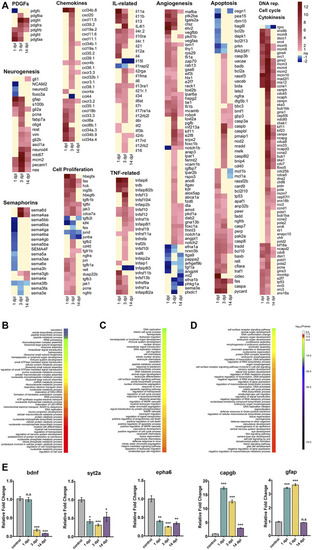
Transcriptome profiling and functional annotation of the telencephalon during early wound healing (1 dpl), early proliferative (3 dpl) and differentiation (14 dpl) stages of zebrafish brain regeneration. |
|
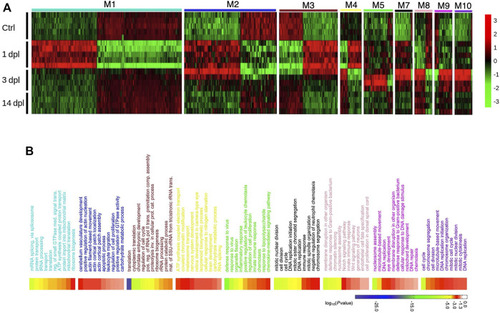
Network analysis of zebrafish brain regeneration at three stages reveals stage-specific modules. |
|
Early wound healing stage (1 dpl) of brain regeneration is similar to brain cancer with respect to induction of metabolism- and neurogenesis-related signaling responses. |
|
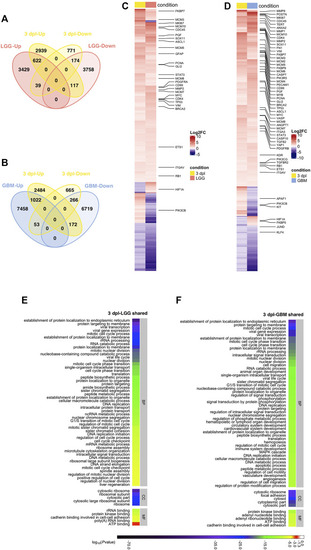
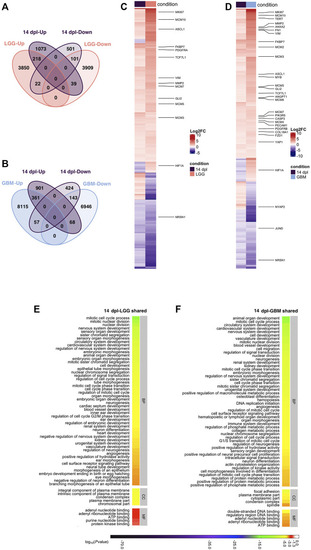
Early proliferative stage (3 dpl) of brain regeneration resembles brain cancer with regard to activation of cell proliferation. (A,B) Venn diagrams showing the number of upregulated (Up) or downregulated (Down) DEGs and the overlap between 3 dpl (yellow) and (A) LGG (pink) and (B) GBM (blue). (C,D) Heatmaps showing the expression of genes shared between 3 dpl (yellow) and (C) LGG (pink) and (D) GBM (blue). Each column represents a condition (3 dpl, LGG or GBM) and each row shows a single gene. The scale bar shows log2 fold changes from high to low regulation, represented by a color gradient from red to purple, respectively. (E,F) DAVID was used to show the most significantly enriched GO-BP (top 50), CC (top 5), and MF (top 5) terms based on transcriptional changes in comparison of 3 dpl with (E) LGG and (F) GBM by using human identifiers of shared DEGs. The heatmap scale shows log10 of the ease p-values for the most significantly enriched GO terms. dpl: days post-lesion, DAVID: database for Annotation, Visualization and Integrated Discovery, GO: Gene Ontology, BP: Biological Process, MF: Molecular Function, CC: Cellular Component, dpl: days post lesion, LGG: low-grade glioma, GBM: glioblastoma. |
|
Differentiation stage (14 dpl) of brain regeneration and brain cancer share mechanisms related to developmental and morphogenetic processes. |
|
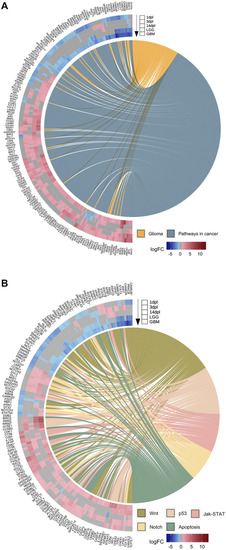
Early wound healing stage of brain regeneration is more similar to LGG and GBM than the proliferation and differentiation stages. |
|
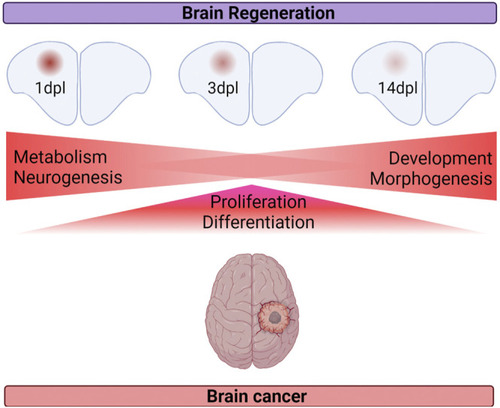
Summary of the shared cellular mechanisms between brain regeneration at three different stages of regeneration and brain cancer. |