- Title
-
Pluripotency factors determine gene expression repertoire at zygotic genome activation
- Authors
- Gao, M., Veil, M., Rosenblatt, M., Riesle, A.J., Gebhard, A., Hass, H., Buryanova, L., Yampolsky, L.Y., Grüning, B., Ulianov, S.V., Timmer, J., Onichtchouk, D.
- Source
- Full text @ Nat. Commun.
|
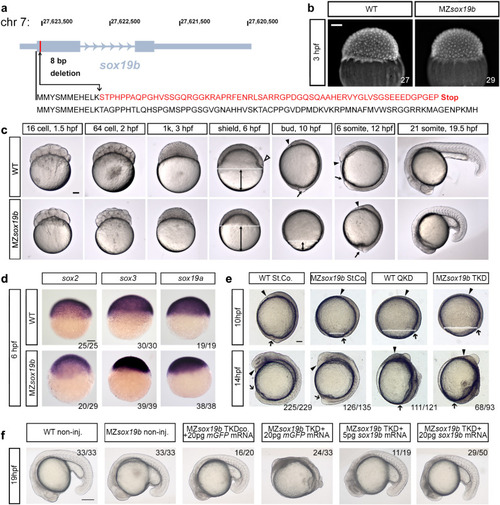
Redundant activity of SoxB1 family factors Sox19b, Sox19a, Sox2 and Sox3 is required at post-gastrulation developmental stages.
a Disruption of the sox19b gene on chromosome 7 by introducing an 8 bp deletion. b No difference in cell division rates between WT and MZsox19b was observed prior to MBT (see also Fig. S1). c MZsox19b embryos are delayed in gastrulation (see Fig. S2 for statistics). Simultaneously collected WT and MZsox19b embryos were let to develop at 28,5 °C, pictures of representative embryos were taken at the indicated time points/developmental stages of the wild type. In zebrafish embryos, embryonic shield forms at 6 hpf at the dorsal side during gastrulation (hollow arrowhead in the wild type). MZsox19b embryos are still phenotypically at 40% epiboly (blastula). Gastrulation ends with tail bud formation at 10 hpf. MZsox19b embryos are still at 80–90% epiboly gastrula stage. d In situ hybridization for sox2, sox3, and sox19a, in WT and MZsox19b embryos, lateral views. e Quadruple Sox19a/b, Sox2, and Sox3 knockdown embryos complete gastrulation, but show later defects in tail bud formation and axis elongation. 1-cell stage wild-type or MZsox19b embryos were injected with control morpholino (StCo), or QKD (quadruple knockdown) mix (Sox2, Sox3, Sox19a, and Sox19b morpholinos), or TKD (triple knockdown, Sox2, Sox3, Sox19a morpholinos), as indicated. f Axis elongation defects in MZsox19b-TKD are rescued by injection of sox19b mRNA. 1-cell stage MZsox19b embryos were injected with either TKD mix or TKDco mix (Sox2, Sox3, Sox19b morpholinos), together with Sox19b or control GFP mRNA, as indicated (see Fig. S3d–g for additional statistics). Double black arrows show the distance from epiboly border (white dotted line) to the vegetal pole. Black arrow—tail bud, black arrowhead—head process. Scale bar 200 µm. Scale bars in b–e: 100 µm, in f: 200 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Maternal Pou5f3 and Sox19b safeguard correct dorso-ventral patterning.
a Comparison of the single mutants, double mutant and wild-type embryos. 10 hpf: MZsox19bspg mutants are arrested in gastrulation similarly to MZspg. Arrow shows abnormally enlarged shield in MZsox19bspg. b, c Double maternal mutants Msox19bspg are dorsalized. b In situ hybridization for dorsal markers noggin1 and Chordin, lateral views, dorsal to the right. Note the circumferential expansion in the Msox19bspg. c Somites (arrowheads) form on the dorsal side in WT, but spread over the Msox19bspg embryo. Dorsal up, anterior to the left. d Normal development of Msox19bspg mutants can be rescued by reducing Chordin, but not Noggin1 levels. The wild-type or Msox19bspg embryos were injected with the indicated morpholinos or non-injected. The numbers show the ratio of embryos with indicated phenotype/ all embryos alive at 22 hpf. The arrows show abnormally expanded blood progenitor cells in the ventralized wild-type embryos. Anterior to the left, dorsal up. e, f Combinatorial (e) and distinct (f) functions of Pou5f3 and SoxB1. e Maternal Sox19b and Pou5f3 safeguard correct D/V patterning. f Pou5f3 is critical for epiboly and gastrulation, redundant action of zygotic SoxB1 (Sox19a, Sox19b, Sox2, and Sox3) is critical for organogenesis. Scale bars in a, b, c: 100 µm, in d: 200 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
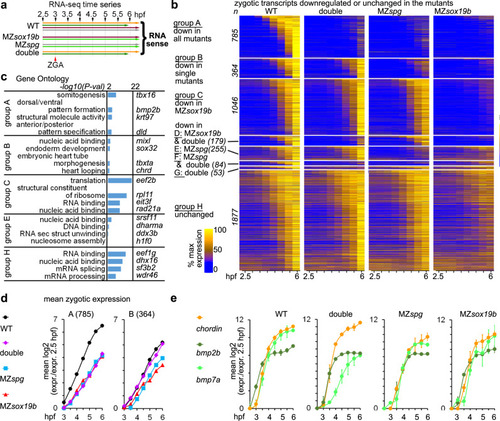
Pou5f3 and Sox19b non-additively activate transcription.
a Experimental setup: RNA-seq time series included eight time points, four replicates for the wild type, and two replicates for each MZspg, MZsox19b, and double mutant MZsox19bspg (see Fig. S4a for the details). Time series, where material was collected in the same experiment, have the same color. b Heatmap of all 4643 zygotic transcripts in the indicated genotypes. 2766 transcripts were downregulated in the mutants (groups A–G),1877 transcripts were not (group H). Zygotic transcripts were sorted by ascending switch time and switch p-value in the wild type. n number of transcripts. c Enrichment in Gene Ontology terms (DAVID). Top four categories per group and example genes are shown. GO: enrichment for “structural molecule activity” in group A was due to the battery of eight keratins, activated by Klf17 in the epithelial layer46. d Non-additive (group A) and compensatory (group B) effects of Pou5f3 and Sox19b on the earliest zygotic transcription. Mean zygotic transcription profiles for the groups A and B, relative to 2.5 hpf. e Chordin/BMPs ratio is increased in the double mutant, but not in the single mutants throughout the time curve. Note that the transcription start of bmp2b and bmp7a, but not Chordin is delayed in the double mutant compared to the WT. In d, e: n(wt) = 4, n(MZspg) = 2, n(MZsox19b) = 2, n(double) = 2, where n is a number of biologically independent experiments. Data are presented as mean values ± SEM. Source data for b, d, and e are provided as a Source Data file. Source data for b–d are provided as Dataset S1. |
|
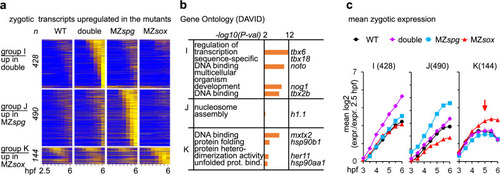
Simultaneous loss of Pou5f3 and Sox19b leads to increase in dorsal gene expression and premature activation of differentiation factors.
a Heatmap for 1062 zygotic transcripts, upregulated in the mutants compared to the wild type. The upregulated transcripts were grouped as indicated in the left (groups I, J, K), and sorted by ascending switch time and switch p-value in the respective mutant. b The groups I, J, K were tested for enrichment in Gene Ontology terms using DAVID. Top four categories per group/example genes are shown. Note that the group I upregulated in MZsox19bspg is enriched for the regulators of transcription and for developmental genes. c Mean zygotic transcription profiles for the groups which are upregulated in the mutants compared to the wild type (I, J, K), relative to 2.5 hpf. Note the early upregulation of the transcripts over the WT in MZsox19bspg (group I) and MZspg (group J), but not in MZsox19b (K), where the transcripts are mostly upregulated at 5 hpf (red arrow). MZsox = MZsox19b. n(wt) = 4, n(MZspg) = 2, n(MZsox19b) = 2, n(double) = 2, where n is a number biologically independent experiments. Data are presented as mean values ± SEM. Source data for a, c are provided as a Source Data file. Source data for a, b are provided as Dataset S2. |
|
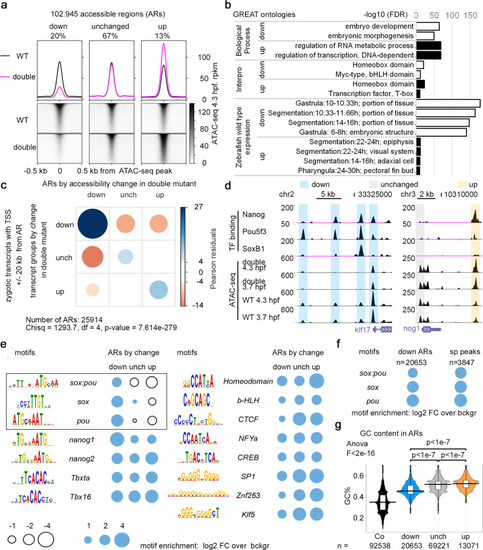
Changes in chromatin accessibility on the cis-regulatory regions underlie expression changes in the double mutants.
a Three groups of accessible regions (ARs), where chromatin accessibility was reduced (“down”), unchanged, or increased (“up”) in MZsox19bspg double mutants relatively to the wild type. Regions were sorted by descending ATAC-seq peak score. b GREAT91 enrichment in Gene Ontology terms for “down” and “up” regions shown in a. Top two categories are shown in “Biological Process” and “Interpro”, four top categories in “Zebrafish wild-type expression”. c “down”, “unchanged” and “up” ARs are enriched around Transcription Start Sites (TSS) of zygotic genes, down, unchanged, and upregulated in MZsox19bspg double mutant, respectively. Chi-square test, p-value is two-tailed. d Pou5f3, SoxB1, and Nanog binding on the cis-regulatory regions of klf17 and nog1 genes, down- or upregulated in MZsox19bspg respectively. UCSC genomic browser view, TF binding and ATAC-seq signal—normalized reads. Note that all three TFs bind to the “down” cis-regulatory regions of klf17, but only Nanog to the “up” cis-regulatory region of nog1. e Enrichment for TF-binding motifs on “down”, “unchanged” and “up” ARs, relative to genomic background. Cognate motifs for Pou5f3 and Sox19b are enriched in “down” and underrepresented in “up” group (black frame). f Enrichment for sox:pou, sox and pou motifs on all “down” regions (ARs down) is close to that on the regions directly bound by Pou5f3 and SoxB1 (sp peaks). g GC nucleotide content of “down” ARs is higher than control genomic regions but lower than in unchanged and “up” ARs. To obtain control regions, genomic coordinates of ARs were shifted 1 kb downstream. Numbers of ARs (n) are indicated below the graphs. The lower border, middle line, and upper border of the white box plots correspond to 0.25, 0.5 (median), and 0.75 quantile, respectively. 1-way ANOVA, p-values in Tukey–Kramer test. Source data for a, b, and d–g are provided as a Dataset S3. Source data for e, f are provided as Dataset S4. Source data for c are provided as Dataset S5. |
|
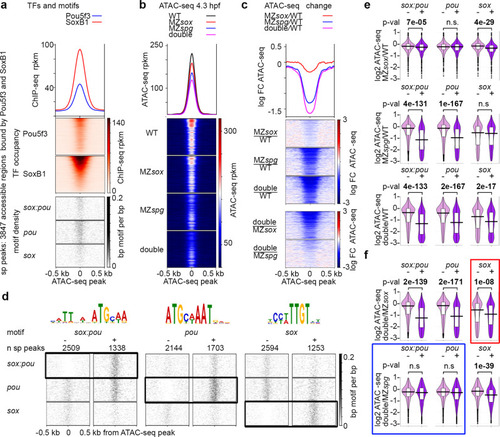
Pou5f3 and Sox19b regulate chromatin accessibility on different motifs.
a–c 3847 accessible regions bound by both Pou5f3 and SoxB1 (sp peaks) were used for analysis. For the heatmaps, the regions were sorted by descending SoxB1 Chip-seq signal. a Heatmaps show TF occupancy and motif density on sp peaks. b Chromatin accessibility on sp peaks is reduced in the single and double mutants at 4.3 hpf. c Chromatin accessibility change. Three top heatmaps: mutant to wild-type change. Two bottom heatmaps: double to single mutant change. Note the strongest chromatin accessibility reduction in the double mutant and the weakest in MZsox19b. d Chromatin accessibility changes at 4.3 hpf were compared between sp peaks without (−) or with (+) motifs indicated above (e, f). Scale-motif density. e Mutants to wild-type change. Sox19b binding to sox motifs and Pou5f3 binding to pou motifs promotes chromatin accessibility. On sox:pou motifs, binding of any factor has a significant effect, but Sox19b effects are weaker. f Double to single mutant change. Blue frame: on sox:pou and pou motifs, chromatin accessibility is similarly reduced in the double MZsox19bspg mutant compared to MZspg. Hence, the gain of chromatin accessibility on sox:pou and pou motifs results from Pou5f3 binding. On some of sox:pou motifs Sox19b may act by facilitating or stabilizing Pou5f3 binding. Red frame: on sox motifs, chromatin accessibility is stronger reduced in MZsox19bspg compared to MZsox19b, suggesting additive or redundant effects of Pou5f3. As shown at Fig. S7g, this effect is due to the sp peaks where both types of motifs (sox and sox:pou or sox and pou) are present. Hence, Pou5f3 and Sox19b may promote chromatin accessibility additively or redundantly when they bind to different motifs within the same AR. MZsox = MZsox19b. At e, f: The lower border, middle line and upper border of the white box plots correspond to 0.25, 0.5 (median), and 0.75 quantile, respectively. n(wt) = 3, n(MZspg) = 3, n(MZsox19b) = 3, n(double) = 4, where n is a number biologically independent ATAC-seq experiments. P-values in two-tailed Student’s t-test. Source data for b, c, e, and f are provided as a Dataset S3. Source data for a, d are provided as Dataset S4. |
|
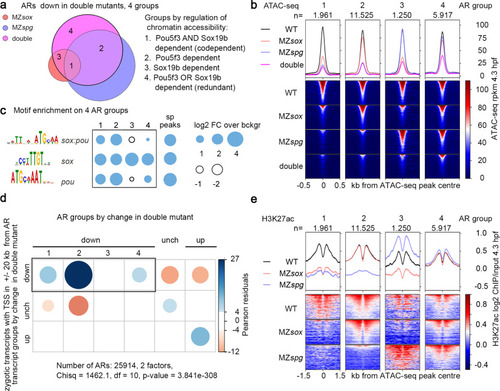
Codependent, Pou5f3-dependent, and redundant cis-regulatory elements regulate the major ZGA wave.
a Venn diagram: 20653 “down” ARs (regions where chromatin accessibility was reduced in the double mutant) were subdivided to groups 1–4 by reduction of chromatin accessibility in the single mutants. b ATAC-seq signals on AR groups 1–4 in the wild type, single and double mutants. ARs were sorted by descending ATAC-seq peak score. c Enrichment for sox:pou, sox and pou motifs in AR groups 1–4 compared to the regions bound by Pou5f3 and SoxB1 (sp peaks). d Chi-squared test. Codependent(1), Pou5f3-dependent (2), and redundant (4) ARs, but not Sox19b-dependent ARs, are enriched around Transcription Start Sites (TSS) of zygotic genes, downregulated in MZsox19bspg double mutant. P-value is two-tailed. e H3K27ac on 4 AR groups depends on the same TFs as accessibility (compare with ATAC-seq in b). (1) codependent ARs: Pou5f3 and Sox19b are required for H3K27ac, (2) Pou5f3-dependent ARs: Pou5f3 is required for H3K27ac, (3) Sox19b-dependent ARs: Sox19b is required for H3K27ac, (4) redundant ARs: any factor is sufficient for H3K27ac, no reduction in the single mutants. ARs were sorted by descending ATAC-seq peak score. MZsox = MZsox19b, double = MZsox19bspg, n number of regions in each group. Source data for a–c, and e are provided as a Dataset S3. Source data for c are provided as Dataset S4. Source data for d are provided as Dataset S5. |
|
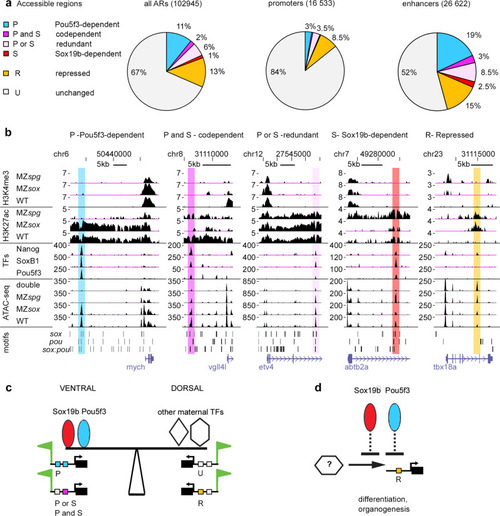
Pou5f3 and Sox19b regulate enhancer activity.
a Pou5f3 and Sox19b regulate chromatin accessibility on more enhancers than promoters. Six types of cis-regulatory elements include four types downregulated in the double mutants (Pou5f3-dependent, codependent, redundant, and Sox19b-dependent), one type upregulated in the double mutant (repressed), and unchanged in the double mutant. Percentages of these types for all ARs, promoters, and enhancers are shown b Examples of enhancer types where chromatin accessibility is directly regulated by Pou5f3 or Sox19b, as indicated (mych, vgll4l, etv4, abtb2a) and indirectly repressed enhancer (tbx18a). UCSC genomic browser view, H3K27ac and H3K4me3 are shown as log2 ChIP/input ratio, ATAC-seq and TF binding as normalized reads, motif occurrence as bars. Note the parallel changes in H3K27ac and chromatin accessibility (for MZspg and MZsox19b). MZsox = MZsox19b, double = MZsox19bspg. Source data for a, b are provided as a Dataset S3. c The model of dorso-ventral balance at ZGA: Pou5f3 and Sox19b prime ventral and ectodermal genes for activation, activating Pou5f3-dependent, codependent and redundant enhancers (“P”, “P and S”, “P or S”). Dorsal and mesendodermal genes are primed by other maternal TFs via independent enhancers (I) and enhancers indirectly repressed by Pou5f3 and Sox19b (“R”, i.e. noggin1 enhancer on Fig.Fig.5d).5d). d Scheme: Pou5f3 and Sox19b indirectly repress premature activation of transcription factors involved in organogenesis and differentiation. |