- Title
-
Insights into the evolution of the ISG15 and UBA7 system
- Authors
- Liu, S., Hu, G., Luo, S., Wu, W., Zhou, Q., Jin, R., Zhang, Y., Ruan, H., Huang, H., Li, H.
- Source
- Full text @ Genomics
|
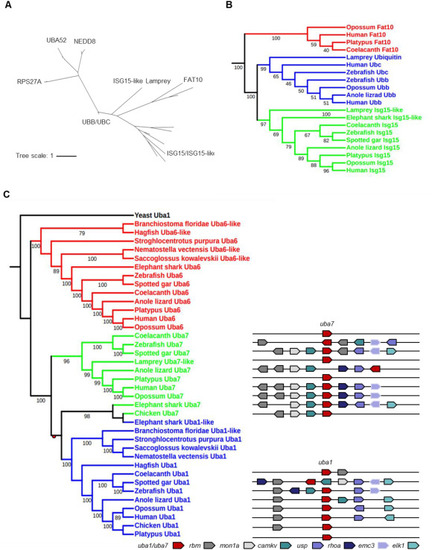
Fig. 1. Phylogeny of ISG15 and UBA7. (A) Phylogenetic tree analyses of the ubiquitin family (ISG15, NEDD8, FAT10, UBA52, RPS27A, and UBB/UBC). The phylogenetic tree was constructed using amino acid multiple alignments generated by ClustalW and the maximum-likelihood method within the MEGA X program. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (5000 replicates) is indicated. (B) Phylogenetic tree analyses of part of the ubiquitin family (ISG15, FAT10, and UBB/UBC). The phylogenetic tree was constructed as described in (A). (C) Phylogenetic (left) and syntenic (right) analyses for Uba1, Uba6, and Uba7. The phylogenetic tree, based on full-length amino acid sequences of Uba1, Uba6, and Uba7, was built using the maximum likelihood method with 5000 bootstrap replicates. Duplication nodes are shown as redcircle. A comparison of genomic synteny between UBA1 and UBA7 is shown on the right, with orthologs of each gene given in the same color. The genes are represented by block arrows whose directions indicate their orientation. |
|
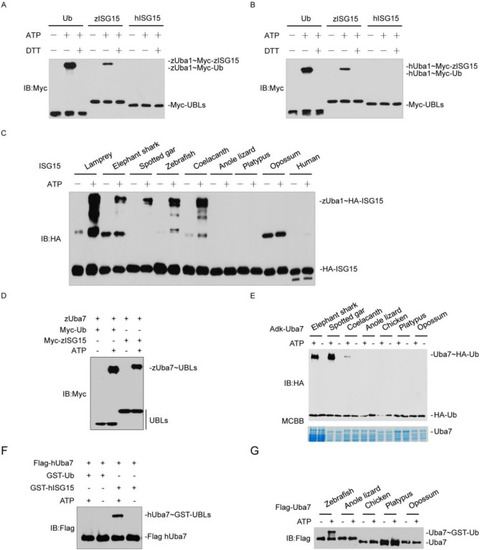
Fig. 2. ISG15-Uba7 from basal vertebrates retain the ancestral Ub-Uba1 phenotype. (A) zUba1 activates Ub, zISG15. The activation was analyzed by immunoblotting with anti-Myc. (B) hUba1 could not activate Ub, zISG15, and hISG15. The activation was analyzed by immunoblotting with anti-Myc. (C) zUba1 activates ISG15s from lamprey, elephant shark, spotted gar, zebrafish. The samples were analyzed by immunoblotting with anti-HA. (D) zUba7 activates Myc-zISG15 and Myc-Ub. The activation was analyzed by immunoblotting with anti-Myc. (E) Activation of Ub by Uba7s from elephant shark, spotted gar, coelacanth expressed in bacteria. The samples were analyzed by immunoblotting with anti-Myc. (F) hUba7 could not activate Myc-hISG15 and Myc-Ub. The activation was analyzed by immunoblotting with anti-Myc. (G) Uba7 proteins of anole lizard, chicken, platypus, and opossum purified from HEK293T cells could not activate Ub. The activation was analyzed by immunoblotting with anti-Flag. |
|
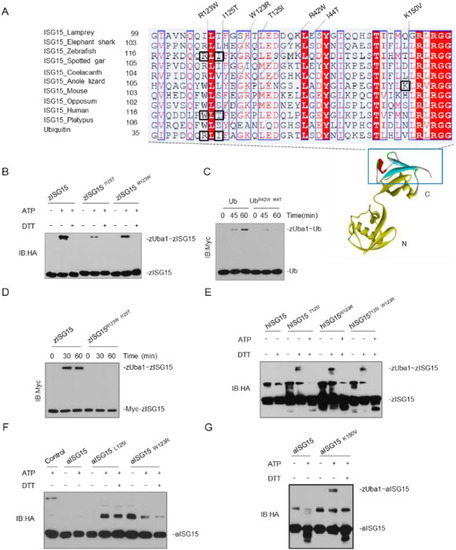
Fig. 3. A determinant in ISG15 contributes to specific ISG15-Uba7 signaling. (A) Alignment of ISG15 sequences from selected species. The multiple alignments were produced using ClustalW. The positions that are important for the properties of ISG15 are highlighted with blue boxes. (B) zUba1 activates Myc-Ub and Myc-UbR123W, I125T. The reactions were prepared at the indicated time points, and immunoblotting was done with anti-Myc. (C, D) zUba1 activates myc-zISG15 and its double-point (C) or single-point (D) mutants; the reactions were prepared at the indicated time points. (E) zUba1 can activate hISG15 T125I, hISG15W123R, and hISG15 I125T, W123R more than wild-type hISG15WT. (F, G) zUba1 activates aISG15 L125I, aISG15W123R (F), and aISG15 K1K150V (G) more than wild-type aISG15WT. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
|
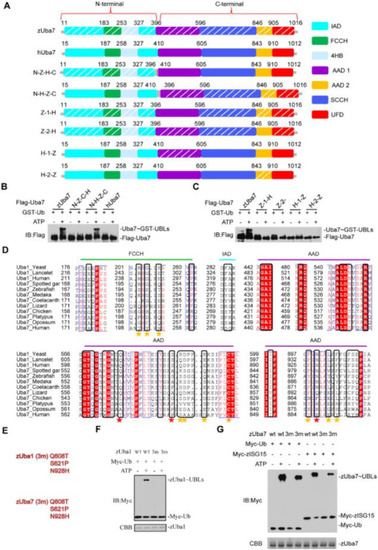
Fig. 4. A determinant in Uba7 contributes to the specific ISG15-Uba7 signaling. (A) Schematic diagram of the N-Z-C-H chimera, containing the N-terminal 396 amino acids of zebrafish, in an otherwise hUba7 background, and N-H-C-Z chimera, which contains the 410 N-terminal amino acids of human in a hUba7 background. The AAD1 and AAD2 domains of zUba7 and hUba7 were interchanged in Z-1-H, Z-2-H, H-1-Z, and H-2-Z. Z-1-H means that the AAD1 domain of zUba7 was replaced by that of hUba7. zAAD1 (396–596), zAAD2 (846–905), hAAD1 (410–605), and hAAD2 (843–910). (B, C) Charging of Ub in vitro by chimeric E1 proteins. Activation was analyzed by immunoblotting with anti-Flag. (D) Sequence alignment of the Ub-interacting regions of Uba1 with Uba7 from the indicated species. Colors indicate amino acid identity and conservation. Residue numbers are indicated to the left, and the domains in which the residues reside are indicated on top of the alignment. Residues involved in contacts with Ub based on crystal structures are indicated by stars under the alignment. (E–G) Mutational analyses of the transthioesterification reaction assays. The corresponding residues for mutation of zUba1 and zUba7 are shown in (E). zUba7 activates Myc-zISG15 and Myc-Ub. Activation was analyzed by immunoblotting with anti-Myc. Wild-type and mutants of zUba1 are shown in (F), whereas wild-type and mutant zUba7 are shown in (G). The amount of zUba1 and zUba7 used in these experiments are shown in the corresponding figure. |
|
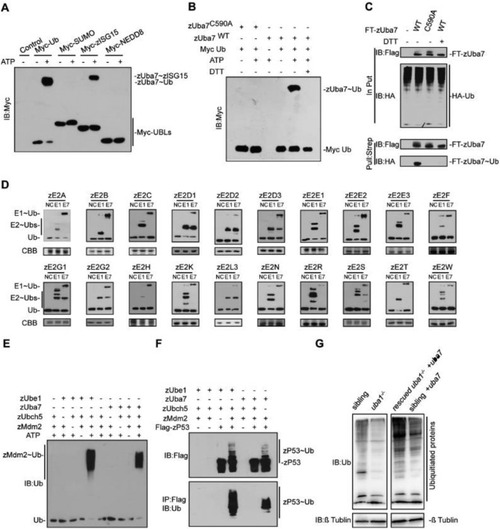
Fig. 5. zUba7 initiates ubiquitination cascades. (A) zUba7 activates Myc-Ub. The activation was analyzed by immunoblotting with anti-Myc. (B) Ub activation by zUba7 requires ATP and Cys 590, and it is sensitive to DTT. (C) Pull down of Flag-Strep-zUba7 (FT-zUba7) and HA-Ub. FT-zUba7 and HA-Ub were expressed in HEK293T cells either alone or together, as indicated. After pull down against the strep tag of zUba7, samples were separated by SDS-PAGE under non-reducing or reducing conditions and were immunoblotted with anti-HA and anti-FLAG antibodies. (D) zUba7 and zUba1 have distinct E2 charging activities in vitro. Assays employed E2s produced in Escherichia coli extracts, HA-Ub and the indicated E1, as described in the Methods. (E) Bacteria expressing Myc-zMdm2 were incubated in the absence or presence of zUba1, zUba7, Ub, and zUbcH5b as indicated. After 1 h at 30 °C, samples were analyzed by immunoblotting with the indicated antibody. (F) In vitro ubiquitination of Flag-zP53 by Myc-zMdm2. (G) Immunoblotting reveals that zUba7 overexpression rescues the Ub level of Zuba1−/−. PHENOTYPE:
|
|
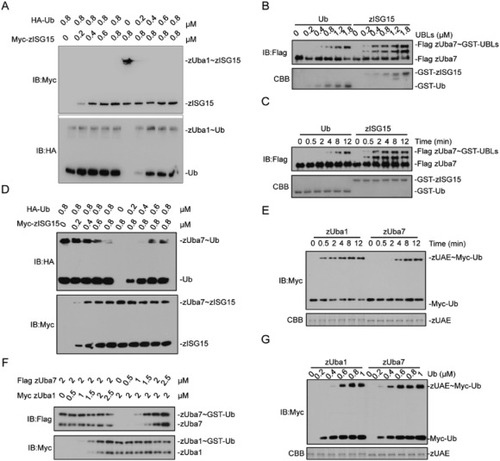
Fig. 6. zUba7 prefers zISG15 and zUba1 prefers Ub. (A), Ub or zISG15 was titrated into reactions containing zUba1, ATP, and zISG15 or Ub fixed at 0.8 μM. (B), GST-Ub and GST-zISG15 titration. zUba7 was incubated with the indicated concentration of GST-UBL for 10 min at 30 °C. Samples were immunoblotted with the anti-Flag. (C), Time course for GST-Ub and GST-zISG15 activation. Reactions containing 1.2 μm GST-UBL and zUba7 were incubated for the indicated time at 30 °C. Samples were immunoblotted with the anti-Flag. (D) Ub or zISG15 was titrated into reactions containing zUba7, ATP, and zISG15 or Ub fixed at 0.8 μM. (E) Kinetics of Ub activation by zUba1 and zUba7. (F) zUba1 or zUba7 was titrated into reactions containing Ub, ATP, and zUba7 or zUba1 fixed at 2 μM. (G) Myc-Ub titration: 2 μM zUba1 or zUba7 was incubated with the indicated concentration of Myc-Ub for 10 min at 30 °C. The samples were immunoblotted with anti-Myc. |