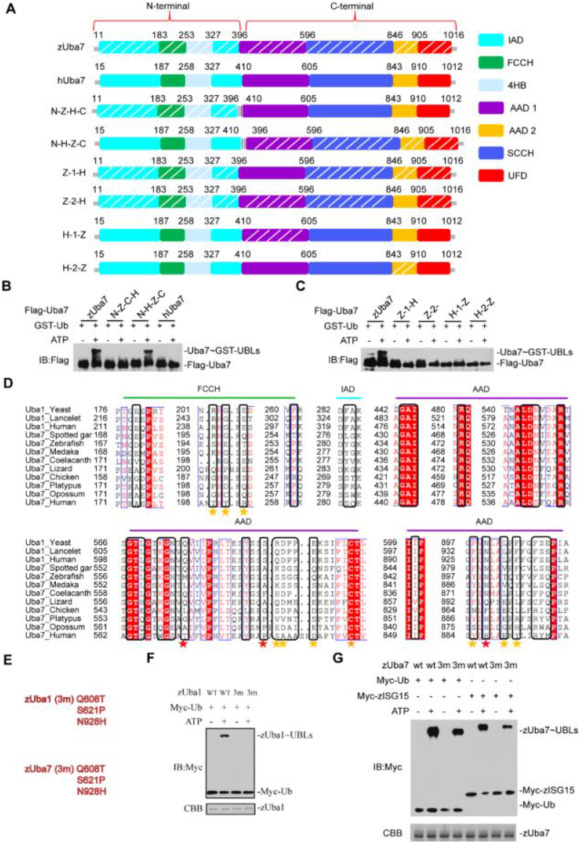
Fig. 4
Fig. 4. A determinant in Uba7 contributes to the specific ISG15-Uba7 signaling. (A) Schematic diagram of the N-Z-C-H chimera, containing the N-terminal 396 amino acids of zebrafish, in an otherwise hUba7 background, and N-H-C-Z chimera, which contains the 410 N-terminal amino acids of human in a hUba7 background. The AAD1 and AAD2 domains of zUba7 and hUba7 were interchanged in Z-1-H, Z-2-H, H-1-Z, and H-2-Z. Z-1-H means that the AAD1 domain of zUba7 was replaced by that of hUba7. zAAD1 (396–596), zAAD2 (846–905), hAAD1 (410–605), and hAAD2 (843–910). (B, C) Charging of Ub in vitro by chimeric E1 proteins. Activation was analyzed by immunoblotting with anti-Flag. (D) Sequence alignment of the Ub-interacting regions of Uba1 with Uba7 from the indicated species. Colors indicate amino acid identity and conservation. Residue numbers are indicated to the left, and the domains in which the residues reside are indicated on top of the alignment. Residues involved in contacts with Ub based on crystal structures are indicated by stars under the alignment. (E–G) Mutational analyses of the transthioesterification reaction assays. The corresponding residues for mutation of zUba1 and zUba7 are shown in (E). zUba7 activates Myc-zISG15 and Myc-Ub. Activation was analyzed by immunoblotting with anti-Myc. Wild-type and mutants of zUba1 are shown in (F), whereas wild-type and mutant zUba7 are shown in (G). The amount of zUba1 and zUba7 used in these experiments are shown in the corresponding figure.