- Title
-
Pipeline for generating stable large genomic deletions in zebrafish, from small domains to whole gene excisions
- Authors
- Tromp, A., Robinson, K., Hall, T.E., Mowry, B., Giacomotto, J.
- Source
- Full text @ G3 (Bethesda)
|
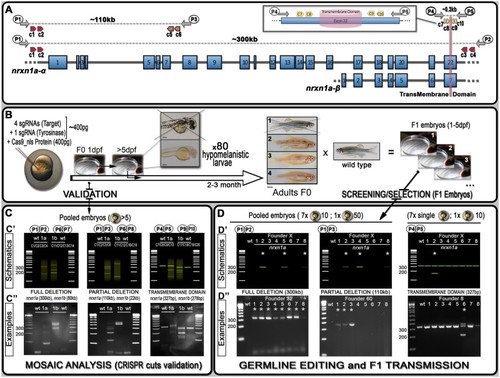
Schematics of neurexin organization and pipeline for generating mutants with large deletions. (A) Representation of neurexin1a (nrxn1a) genomic region together with sgRNA (CRISPR) target sites (c1–10) as well as primers used for validation and screening. The size of exons, intron gaps, and protein domains (and compared with one another) are not drawn to scale. (B) Pipeline designed to generate large deletions in zebrafish; 24 h post-injection, pools of five embryos are collected to evaluate the efficiency of the Cas9/sgRNAs mixes (see (C) mosaic analysis). Following validation, 80× 5dpf-larvae are selected based on their pigmentation phenotype. Once adult, F0 animals are crossed against wild-type to identify carriers/founders able to transmit the designed deletion to F1 mutants (see (D) stable validation). (C) To analyze/validate the efficiency of our sgRNAs-mixes, we proceeded with a basic PCR approach. Depending on the size of the target, a wild-type amplicon may be amplified; with the presence of a wild-type product hampering the amplification/detection of DNA remodeling (especially if rare). If large DNA deletion(s) happens, one should observe smearing with the presence of clear amplicon(s), as presented in the C’ schematics. The lower panel (C”) present the results we obtained for nrxn1a and nrxn1b transient validations. These transient validations can also be performed on individual embryos. (D) The same PCR approach is conducted to identify F0 adult founders. This time, eight batches are collected as schematically described. As opposed to the mosaic analysis (transient approach), a sharp band(s) should be observed, evidencing the presence of a large deletion. Upper panels (D’) represent theoretical results for one specific founder, while lower panels (D”) present PCR profiles obtained during our screen against the different nrxn1a deletion design. Founder frequency was evaluated at approximately 1/10. Asterisks* highlight positive founders carrying the desired mutation. All sequencing data as well as alignments and annotated gene files are available in the Supplementary zip file S04 with mutations summarized in Supplementary Table S04. A step-by-step protocol is also available in Supplementary file S05. |
|
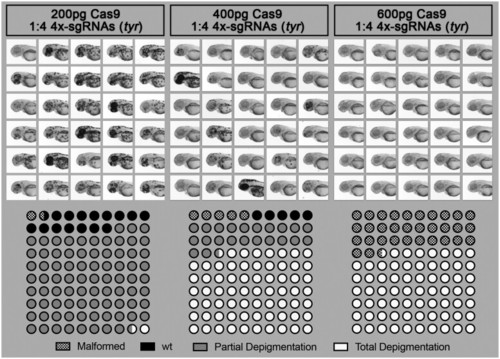
Effect of Cas9/sgRNA concentration on lethality, cutting efficiency, and phenotype outcome in zebrafish. Different concentrations of Cas9 protein mixed with 4× sgRNA guides against the gene tyrosinase (tyr) were injected into the yolk of one-cell stage zebrafish embryos. Injected animals were then observed at 2 dpf to score toxicity (malformation or death), partial depigmentation, or total depigmentation (suggesting complete bi-allelic tyr-knockout). A mix of 400 pg of Cas9/sgRNAs was selected to be used routinely in our pipeline. |
|
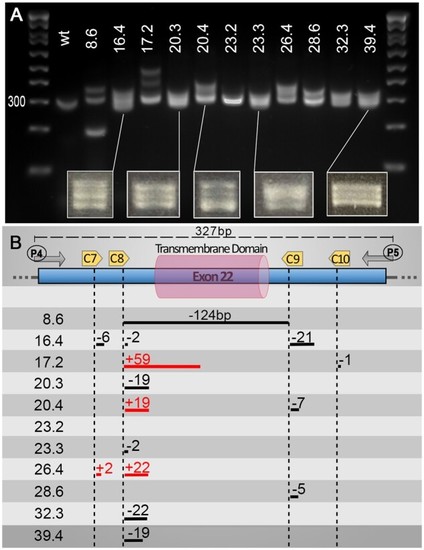
Heteroduplexes can help identify small indels using basic PCR screening approaches. Although not the primary aim of our pipeline, we found that, using a PCR screening approach, one can also easily detect “traditional” small indels due to an aberrant migration of heteroduplex PCR products. We found that at least a 2 bp difference is sufficient to trigger a significant slower migration of potential heteroduplexes. We found that the heteroduplexes were best observed with an amplicon size of around 300 bp, allowing clear visualization of two or three bands on the gels. Additionally, most of our targets/cuts did not sit in the center of the PCR amplicon, which might be a factor in promoting the formation of such heteroduplexes. (A) Gel presenting migration of PCR amplicons using primers P4 and P5 on validated F1 animals (Number X.x, i.e. 8.6, corresponds to fish number followed by embryo sample number). (B) Schematic representation of mutations found in the different animals presented in the upper panel. Distances are approximate only (not drawn to scale). Associated sequencing data are available in Supplementary zip file S07. |