Figure 3
- ID
- ZDB-FIG-211216-162
- Publication
- Tromp et al., 2021 - Pipeline for generating stable large genomic deletions in zebrafish, from small domains to whole gene excisions
- Other Figures
- All Figure Page
- Back to All Figure Page
|
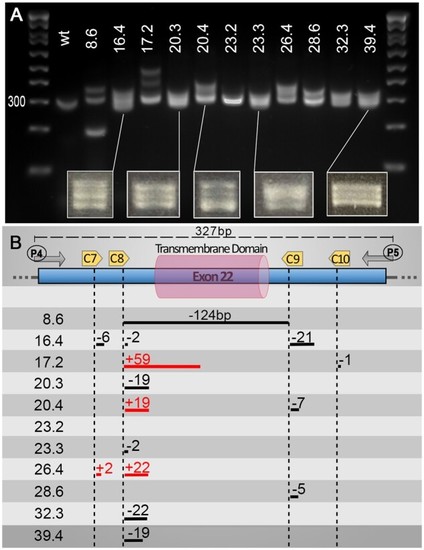
Heteroduplexes can help identify small indels using basic PCR screening approaches. Although not the primary aim of our pipeline, we found that, using a PCR screening approach, one can also easily detect “traditional” small indels due to an aberrant migration of heteroduplex PCR products. We found that at least a 2 bp difference is sufficient to trigger a significant slower migration of potential heteroduplexes. We found that the heteroduplexes were best observed with an amplicon size of around 300 bp, allowing clear visualization of two or three bands on the gels. Additionally, most of our targets/cuts did not sit in the center of the PCR amplicon, which might be a factor in promoting the formation of such heteroduplexes. (A) Gel presenting migration of PCR amplicons using primers P4 and P5 on validated F1 animals (Number X.x, i.e. 8.6, corresponds to fish number followed by embryo sample number). (B) Schematic representation of mutations found in the different animals presented in the upper panel. Distances are approximate only (not drawn to scale). Associated sequencing data are available in Supplementary zip file S07. |