- Title
-
Y705 and S727 are required for mitochondrial import and transcriptional activities of STAT3 and regulate proliferation of embryonic and tissue stem cells
- Authors
- Peron, M., Dinarello, A., Meneghetti, G., Martorano, L., Betto, R.M., Facchinello, N., Tesoriere, A., Tiso, N., Martello, G., Argenton, F.
- Source
- Full text @ Development
|
|
|
|
|
|
|
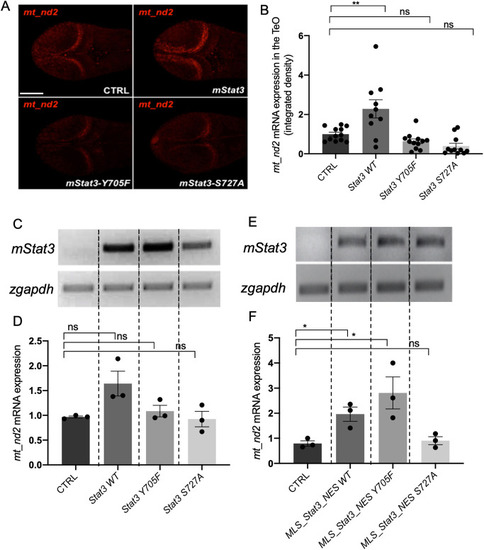
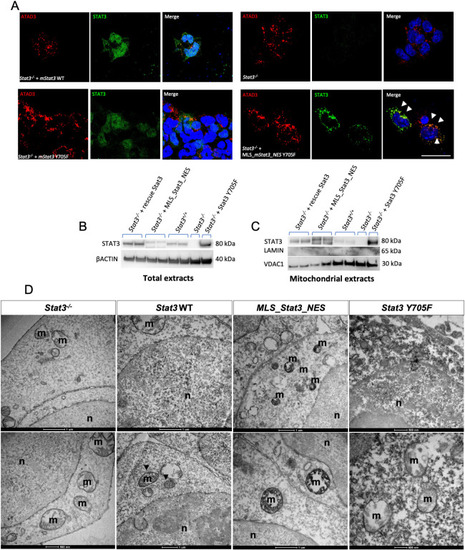
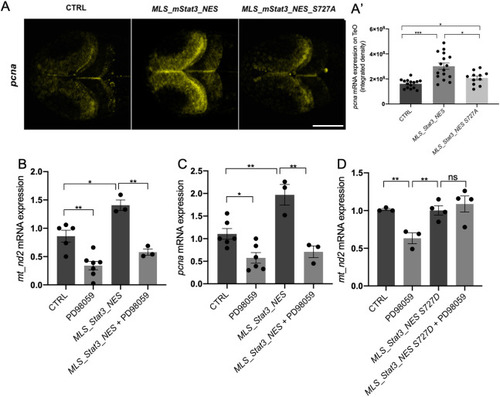
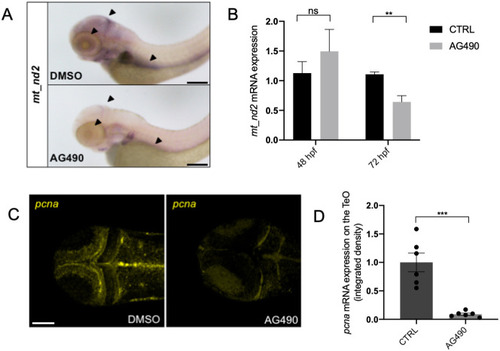
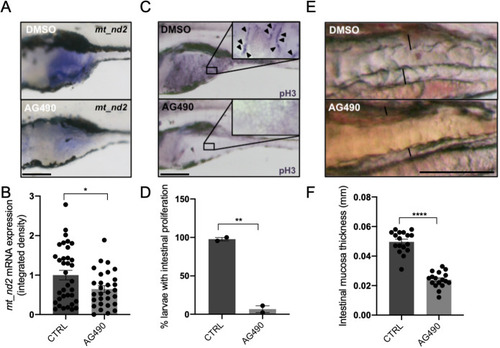
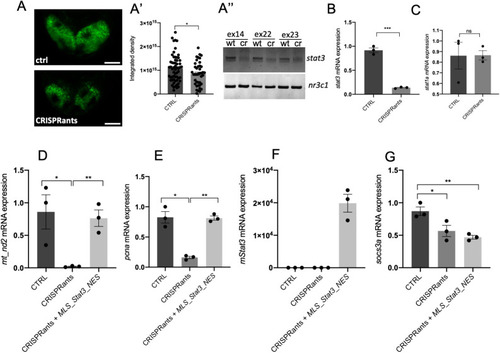
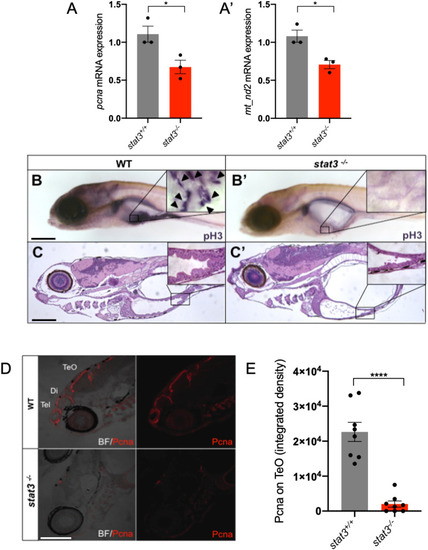
mitoSTAT3 transcriptional activity relies on both S727 and Y705 phosphorylation. (A) FISH with mt_nd2 probe in the TeO of 48 hpf embryos injected with mRNA encoding the indicated mutated forms of mStat3. (B) Fluorescence quantification of mt_nd2 mRNA expression in the TeO (n=10). (C) RT-PCR analysis of mStat3 transcripts detected at 48 hpf in embryos injected with the indicated form of mStat3 mRNA; zgapdh was used as internal control. (D) qRT-PCR analysis of mt_nd2 transcript levels at 48 hpf normalised to zgapdh. (E) RT-PCR analysis of MLS_mStat3_NES transcripts detected at 48 hpf in embryos injected with indicated form of mitochondria-targeted mStat3 mRNA; zgapdh was used as internal control. (F) qRT-PCR analysis of mt_nd2 transcript levels at 48 hpf normalised to zgapdh. Data are mean±s.e.m. *P<0.05; **P<0.01 (unpaired two-tailed t-test on three independent biological samples, where n not specified). ns, not significant. Scale bar: 100 μm. |
|
|
|
|
|
|
|
|
|
|
|
|