- Title
-
Loss of vacuolar-type H+-ATPase induces caspase-independent necrosis-like death of hair cells in zebrafish neuromasts
- Authors
- Santra, P., Amack, J.D.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
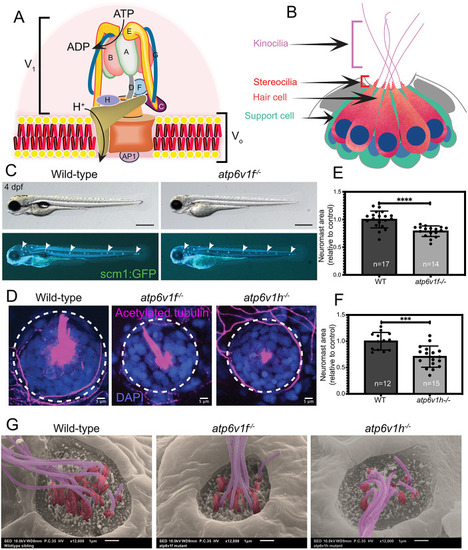
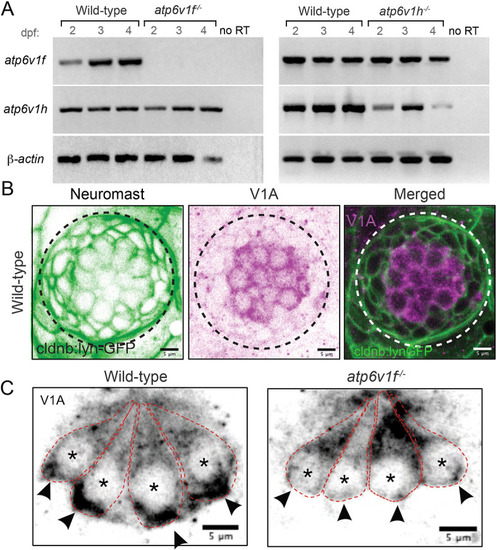
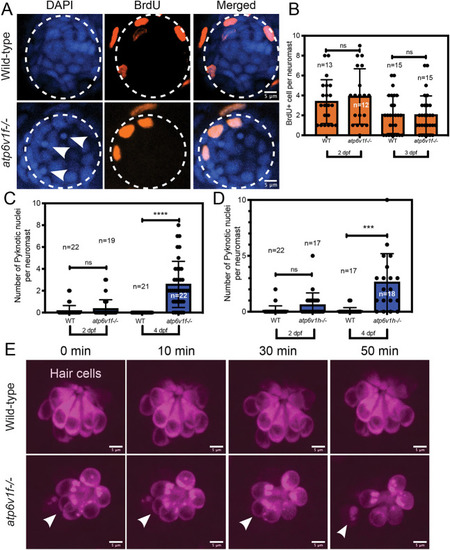
(A) Reverse transcriptase (RT)-PCR analysis of total mRNA from wild-type and V-ATPase mutant embryos at 2, 3 and 4 dpf. atp6v1f mRNA is not detected in atp6v1f−/− embryos, whereas atp6v1h mRNA is detected in the atp6v1h−/− embryos. β-actin mRNA was amplified as a positive control, and reactions without reverse transcriptase (no RT) were negative controls. (B) Antibodies against the V-ATPase V1A subunit (magenta) show enriched staining in centrally localized hair cells in a wild-type neuromast labeled by Tg(cldnb:lynEGFP) expression (green). Dashed line circles indicate the neuromast boundary. (C) Optical sections of hair cells reveal that the V1A subunit localizes throughout wild-type hair cells with an accumulation in the basal region. This basal localization is disrupted in atp6v1f−/− mutant hair cells. Approximate boundaries of individual hair cells are outlined, and asterisks mark hair cell nuclei. Arrowheads indicate basal accumulation of V1A in wild-type hair cells, and lack thereof in atp6v1f−/− hair cells. |
|
|
|
|
|
|
|
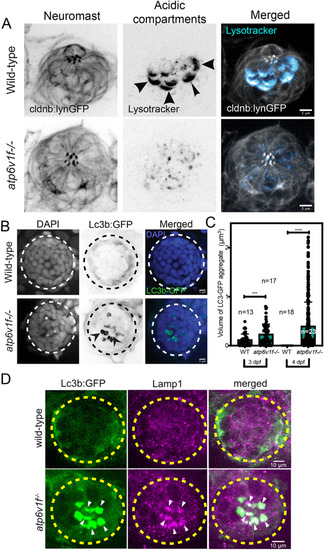
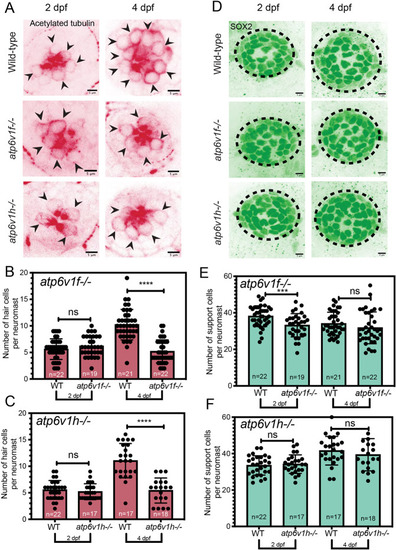
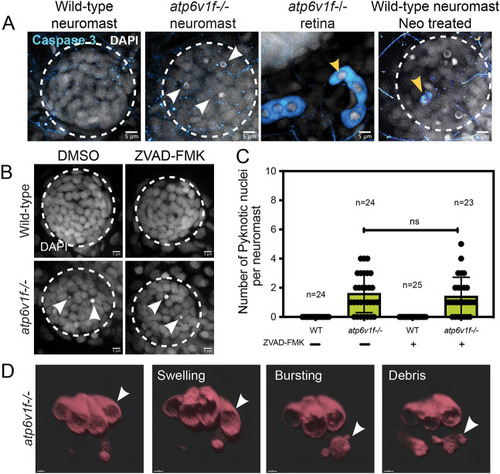
(A) Cleaved Caspase 3 staining was absent in wild-type and atp6v1f−/− mutant neuromasts at 4 dpf, but was detected in the retina of atp6v1f−/− embryos, and in wild-type hair cells treated with neomycin (Neo) that induces Caspase 3 activation (yellow arrowheads). Nuclei were detected using DAPI. White arrowheads indicate pyknotic nuclei. (B) DAPI staining of neuromast nuclei at 4 dpf in wild-type and atp6v1f−/− embryos treated with either DMSO (control) or 300 µM ZVAD-FMK from 2 dpf to 4 dpf. White arrowheads indicate pyknotic nuclei. Dashed line circles indicate the neuromast boundary. Scale bars: 5 µm. (C) Quantification of pyknotic nuclei per neuromast in control and 300 µM ZVAD-FMK-treated embryos. n=number of embryos. ns, not significant by unpaired Student's t-test with Welch's correction. (D) 3D rendering of time-lapse snapshots of Tg(myo6b:tdtomato); atp6v1f−/− hair cells undergoing necrosis-like morphological changes. The white arrowhead follows one hair cell swelling and then bursting over time. Scale bars: 3 µm. |
|
|