- Title
-
Crk adaptor proteins are necessary for the development of the zebrafish retina
- Authors
- Stergas, H.R., Kalbag, Z., St Clair, R.M., Talbot, J.C., Ballif, B.A., Ebert, A.M.
- Source
- Full text @ Dev. Dyn.
|
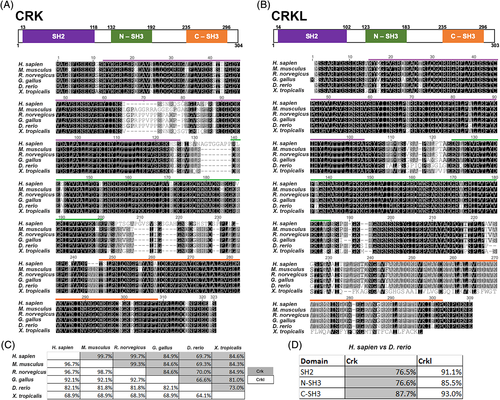
CRK and CRKL are conserved across vertebrates, including Danio rerio. A and B, Multiple protein sequence alignments of CRK (A) and CRKL (B) performed using the Geneious R10 software. Alignment of Homo sapien, Mus musculus, Rattus norvegicus, Gallus gallus, Danio rerio, and Xenopus tropicalis amino acid sequences colored for similarity in greyscale. Domain structure is modeled above alignments (H. sapien amino acid numbers), and functional domains are color coded in both the diagram and highlighted above in the sequence alignment: SH2 domain (purple), N-SH3 domain (green), C-SH3 domain (orange). C, Table showing percent identity of protein sequences compared between each species. Grey boxes correspond to CRK percent identity and white boxes correspond to CRKL. D, Comparison of H. sapien and D. rerio percent identical amino acids of each functional domain. Grey boxes correspond to CRK percent identity and white boxes correspond to CRKL |
|
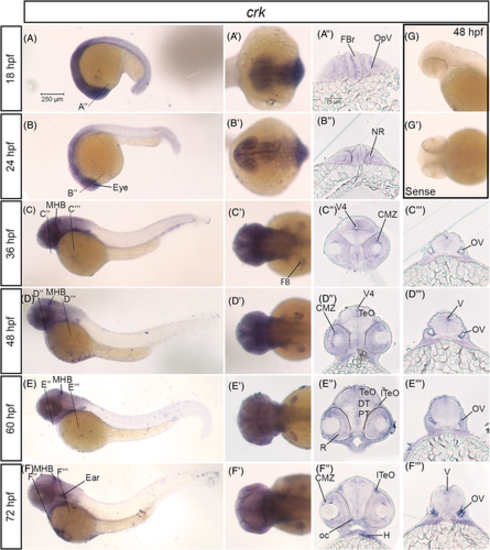
crk mRNA is expressed in the developing nervous system in zebrafish. A–F, Lateral whole mount and (A′–F′) dorsal whole mount in situ hybridization embryos for crk expression at stages 18 hpf–72 hpf. Twenty micron transverse sections through the forebrain (A″–F″) and hindbrain (C″–F″) of in situ embryos at the lines indicated in (A–F). G, Lateral and (G′) dorsal images of sense control probe on 48 hpf embryos. Scale bar at 250 μm (A) and 75 μm (A″). CMZ, ciliary marginal zone; DT, dorsal thalamus; FB, fin bud; FBr, forebrain; H, heart; lTeO, lateral optic tectum; MHB, midbrain-hindbrain boundary; NR, neural retina; oc, optic chiasm; OpV, optic vesicle; OV, otic vesicle; PT, posterior tuberculum; R, retina; TeO, optic tectum; V, ventricle; V4, fourth ventricle EXPRESSION / LABELING:
|
|
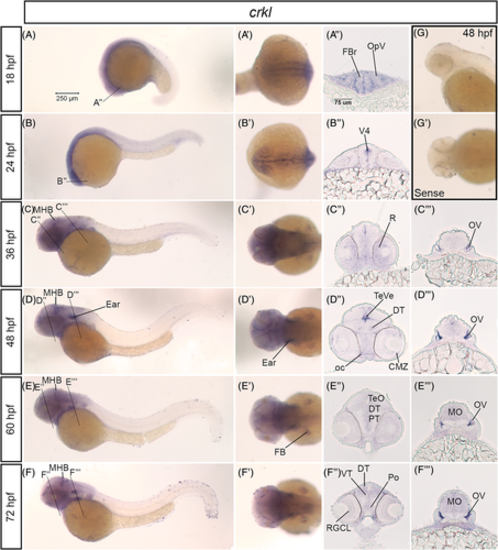
crkl mRNA is expressed in the developing nervous system in zebrafish. A–F, Lateral whole mount and (A′–F′) dorsal whole mount images of in situ hybridization embryos for crkl expression at 18 hpf–72 hpf stages. Twenty micron transverse sections through the forebrain (A″–F″) and hindbrain (C″–F″) at the lines indicated in (A–F). G, Lateral and (G′) dorsal images of sense control probe on 48 hpf embryos. Scale bar at 250 μm (A) and 75 μm (A″). CMZ, ciliary marginal zone; DT, dorsal thalamus; FB, fin bud; FBr, forebrain; MBH, midbrain-hindbrain boundary; MO, medulla; oc, optic chiasm; OpV, optic vesicle; V4, fourth ventricle; OV, otic vesicle; Po, preoptic region; PT, posterior tuberculum; R, retina; RGCL, retinal ganglion cell layer; TeVe, tectal ventricle; TeO, optic tectum; VT, ventral thalamus EXPRESSION / LABELING:
|
|
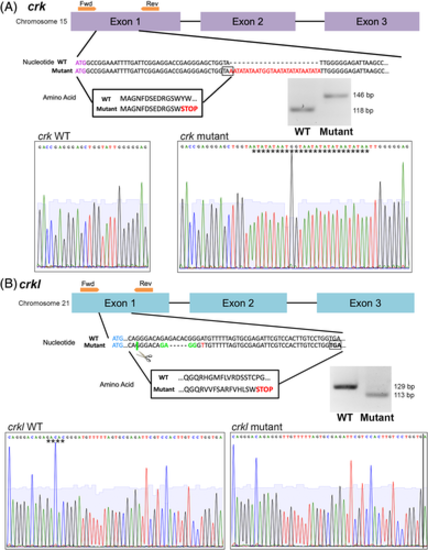
crk and crkl mutant zebrafish lines generated by CRISPR/Cas9. Exon structure, nucleotide sequence, and amino acid sequence diagram of mutations generated by CRISPR/Cas9. Sanger sequencing reads of wild-type and mutant alleles, inserted (crk) or deleted (crkl) sequence denoted by stars. Agarose gel images of wild-type and mutant alleles after crk and crkl genotyping PCRs and, for crkl, the PCR product is digested with MnlI enzyme. A, crk mutant allele. Exons 1–3 of crk shown in purple, forward and reverse primers for genotyping shown in orange on exon structure diagram. crk start codon shown in purple lettering, 28 bp insertion mutation shown in red lettering in the nucleotide sequence with predicted STOP codon boxed. Resulting amino acid sequence boxed. B, crkl mutant allele. Exons 1–3 of crkl diagrammed in blue, forward and reverse primers around the mutation site shown in orange above exon 1. crkl start codon shown in blue lettering, 5 bp deletion site shown with dash marks in mutant allele, MnlI enzyme recognition and cut site for genotyping marked with green, point mutation marked with red lettering and predicted STOP codon boxed. Resulting amino acid sequence boxed |
|
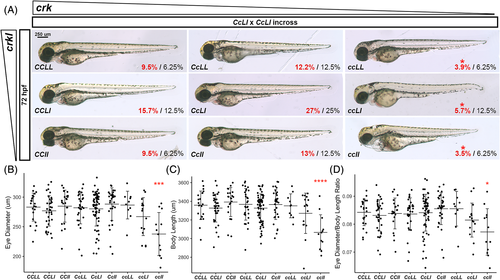
crk and crkl are necessary for proper embryonic development and proper eye size in zebrafish. A, Brightfield whole mount images of 72 hpf embryos resulting from crk heterozygous/crkl heterozygous (CcLl) incross. Genotype of each representative embryo is shown on bottom left corner of the image: C, wild-type crk allele; c, mutant crk allele; L, wild-type crkl allele; l, mutant crkl allele. Bottom right corner of each image shows the actual percentage (red) over expected percentage (black) of embryos of that genotype randomly selected and successfully genotyped from 230 embryos (N = 9). Red star above cc genotype actual percentages denotes unequal proportions. Scale bar at 250 μm in CCLL image. B, Graph of eye diameter; C, body length; and D eye diameter normalized over body length for each genotype analyzed by one-way ANOVA and Tukey multiple comparison test (eye diameter ANOVA P = .000043, CCLL vs. ccll P = .00022; body length ANOVA P = .000005, CCLL vs. ccll P = .000022; eye diameter normalized to body length ANOVA P = .00962, CCLL vs. ccll P = .0307; N = 10, n = 230) PHENOTYPE:
|
|
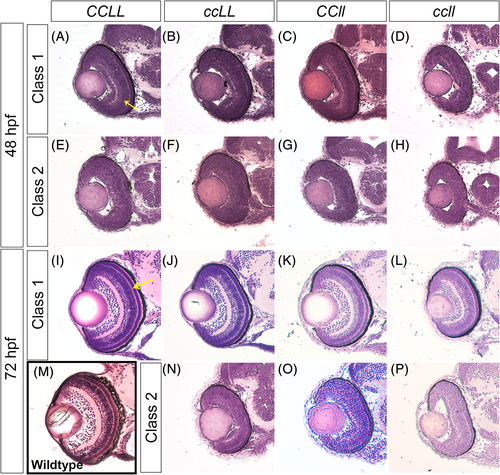
Loss of Crk adaptor proteins leads to retinal lamination defects. Transverse sections of zebrafish embryos stained with Hematoxylin and Eosin through the center of the forebrain, including the retina, of CCLL (A, E, I), ccLL (B, F, J, N), CCll (C G, K, O), and ccll (D, H, L, P) at 48 hpf (A–H) and 72 hpf (I–P). For wild-type comparison, retina section of a 72 hpf Sofa1 transgenic embryo (M),30 the genetic background of the mutant lines. Embryos were classified by retinal lamination phenotype based on % inner plexiform layer (IPL) per total outer curve (48 hpf) and % outer plexiform layer (OPL) per total outer curve (72 hpf) measurements (see Figure 7 for quantification, yellow arrows in A and I indicate IPL and OPL in 48 hpf and 72 hpf, respectively), and sample images of each class are shown: 48 hpf class 1 (A–D), 48 hpf class 2 (E–H), 72 hpf class 1 (I–L), and 72 hpf class 2 (N–P) PHENOTYPE:
|
|
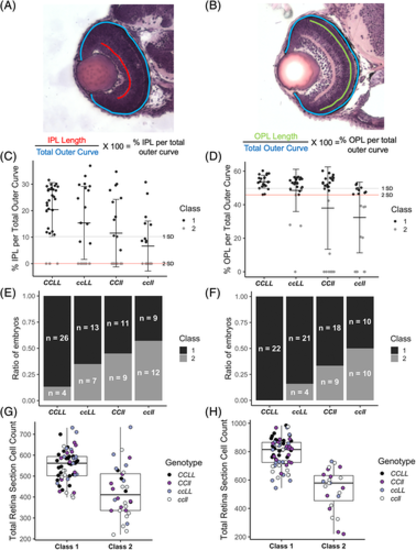
Retinal lamination phenotypes in Crk adaptor mutant embryos separate into two classes and loss of Crk adaptors leads to fewer cells in the retina. Quantifications of the inner retina phenotype of 48 hpf and 72 hpf CCLL, ccLL, CCll, and ccll transverse sections from Figure 6. A and B, Schematics depicting lamination defect quantification to generate the % IPL per total outer curve and % OPL per total outer curve respectively. C and D, Lamination classification of Crk mutant embryos based on retina section % inner plexiform layer (IPL) per total outer curve (C, 48 hpf) and % outer plexiform layer (OPL) per total outer curve (D, 72 hpf). Gray line indicates one SD below the CCLL mean and red line indicates two SDs below the mean, which is the classification cutoff. Class 1 embryos (black dots) have % plexiform layer measurements above the red line and class 2 embryos (gray dots) have % plexiform layer measurements below the red line. Error bars represent SD. E and F, Percent of embryos in each genotype that fall into class 1 and class 2 for lamination phenotype. E, 48 hpf; F, 72 hpf. Black bar indicates class 1, gray bar indicates class 2. Sample size for each genotype and class is denoted in each bar. G and H, Box plots showing total number of cells in each retina section by lamination class (class 1/class 2) separated by genotype. G, 48 hpf; H, 72 hpf. Black dots, CCLL; dark purple dots, ccLL; light purple dots, CCll; white dots, ccll. C, wild-type crk allele; c, mutant crk allele; L, wild-type crkl allele; l, mutant crkl allele |