- Title
-
Nuclear exosome HMGB3 secreted by nasopharyngeal carcinoma cells promotes tumour metastasis by inducing angiogenesis
- Authors
- Zhang, K., Liu, D., Zhao, J., Shi, S., He, X., Da, P., You, Y., You, B.
- Source
- Full text @ Cell Death Dis.
|
|
|
Representative IHC images of extracellular HMGB3 staining and the red arrow points to part of the extracellular staining area. B Statistical comparison of extracellular HMGB3 score in TNM stages. C Statistical comparison of extracellular HMGB3 score in the two groups with or without metastasis. D The pan-cancer analysis of the chromosome aneuploidy. E Statistical comparison of the MN numbers in TNM stages. F Statistical comparison of the MN numbers in the two groups with or without metastasis. G Spearman correlation between nuclear HMGB3 score and extracellular HMGB3 score. H Spearman correlation between nuclear HMGB3 score and the MN numbers. Pearson correlation coefficient (r2) and P value are shown. Mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, student’s test. |
|
|
|
|
|
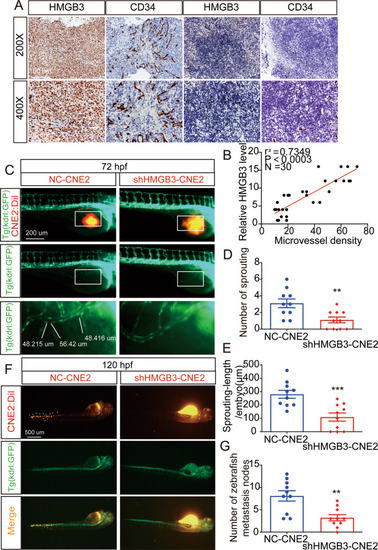
A Representative images of IHC for CD34 and HMGB3 in NPC tissues. The scale bar in 200× images represents 100 µm. B Spearman correlation between HMGB3 score and MVD in NPC patients. Pearson correlation coefficient (r2) and P value are shown. C The overall shape of 96 hpf embryos after injection of about 10 nL of Matrigel/CNE2-NC or Matrigel/CNE2-shHMGB3 solution in the perivitelline space at 48 hpf. White boxes indicate regions of pictures shown in the bottom two pictures and the analysis of the subintestinal venous plexus (SIV) was conducted. The scale bar represents 200 µm. D Quantifications with the number of sprouting of ten embryos per group. E Quantifications with the sprouting-length per embryo of ten embryos per group. F At 120 hpf, the migration of CNE2 cells was measured using fluorescence microscopy. The scale bar represents 500 µm. G Quantification of migratory cell numbers of ten embryos per group. Mean ± SD, **P < 0.01, ***P < 0.001, student’s test. PHENOTYPE:
|
|
|
|
|