- Title
-
Phenotypically Anchored mRNA and miRNA Expression Profiling in Zebrafish Reveals Flame Retardant Chemical Toxicity Networks
- Authors
- Dasgupta, S., Dunham, C.L., Truong, L., Simonich, M.T., Sullivan, C.M., Tanguay, R.L.
- Source
- Full text @ Front Cell Dev Biol
|
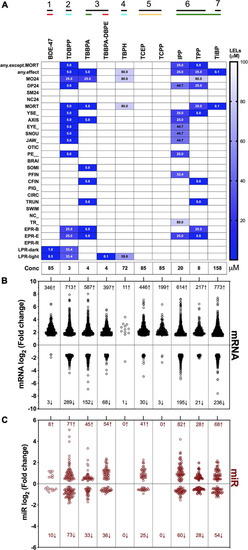
Chemical name, structure, abbreviation, CAS number, physicochemical classes, EC80 s and exposure concentrations (“Conc”) of flame retardant chemicals (FRCs) used in this study. “Conc” represents concentration used for exposures in the study. For FRCs, EC80s [exposure concentrations demonstrating 80% morphological effects based on |
|
Phenotypic screening, mRNA sequencing and miR sequencing of 10 FRCs. |
|
Major gene ontology (GO) processes across FRCs based on |
|
Heatmap representing log2 fold changes of (A) top mRNAs and (B) top miRs across FRCs. Up to 5 genes with highest increase and decrease in mRNA or miR expression levels were selected and their fold changes were plotted for all FRCs. Each column dendrogram color represents a cluster. |
|
mRNA-miR interactions across FRCs based on experimentally validated predictions. (A) FRC-mRNA-miR co-regulatory network based on TFmiR gene-miR interactions. Only mRNA-miR pairs with reverse expression directions were considered for the network. (Red) FRCs, (Blue) miRs, (Yellow) miRs that were differentially expressed across 4 or more FRCs, with larger nodes denoting higher number of FRCs. mRNAs are represented within the connected lines. (B) Selected miRs that were decreased across multiple FRCs, with fold changes and major functions (based on GO analysis) of their anti-correlated mRNA targets in specific FRCs. Numbers represent FRC classes based on Figure 1. TBPH and TCPP not represented since there were no miR disruptions. |
|
Comparison of TBBPA-DBPE and TCEP for neurotoxic effects. TBBPA-DBPE exposures showed only LPR phenotypes while TCEP showed no phenotype. |
|
mRNA-transcription factor (TF) interactions across FRCs according to experimentally validated predictions. |
|
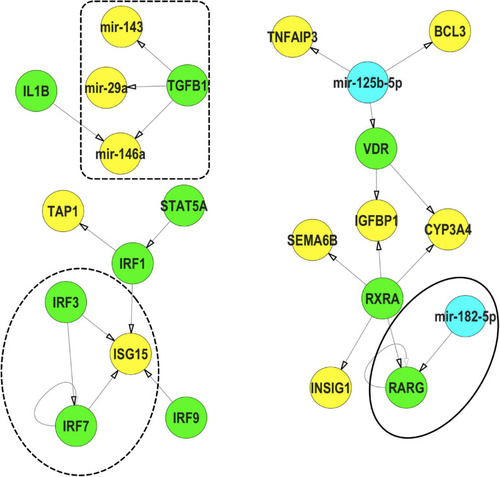
Representative interactions between mRNA, miR and TFs that show a feed forward loop (FFL, dotted circle), a TF regulating a miR (dotted box) and a miR regulating a TF (solid circle) within IPP exposures. (Green) TFs, (Blue) miRs, (Yellow) mRNA. These interactions were selected from the mRNA-miR-TF co-regulatory network for IPP. All interaction data based on TFmiR experimentally validated predictions. |
|
mRNA expression for TBBPA vs its derivative, TBBPA-DBPE. |
|
mRNA expression for BDE-47, aryl phosphates and chlorinated phosphates. (A,C) Heatmaps representing log2 fold changes of mRNA expression; colored bars on the rows represent mRNA clusters. Numbers within bars represent cluster numbers for specific mRNA sets. (B,D) GO processes for unique mRNAs that were increased or decreased uniquely in various clusters. |