- Title
-
Retinal Pigment Epithelium and Neural Retinal Progenitors Interact Via Semaphorin 6D to Facilitate Optic Cup Morphogenesis
- Authors
- Cechmanek, P.B., Hehr, C.L., McFarlane, S.
- Source
- Full text @ eNeuro
|
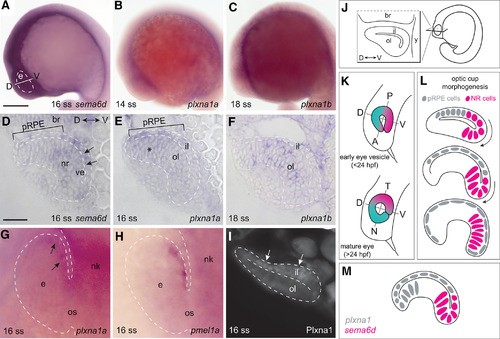
EXPRESSION / LABELING:
|
|
|
|
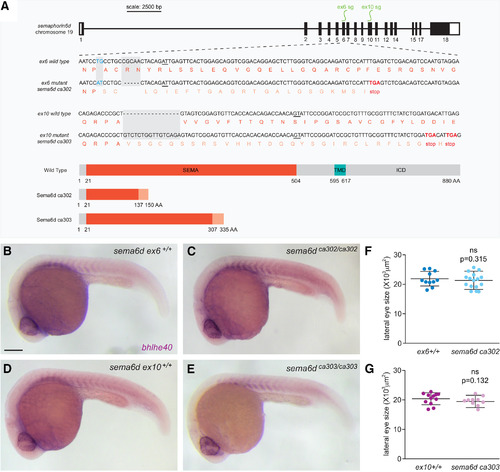
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
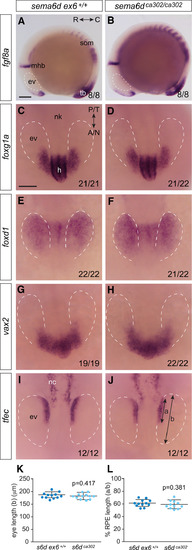
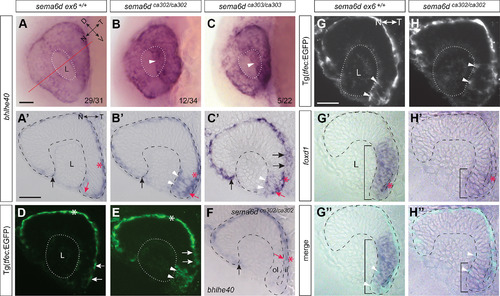
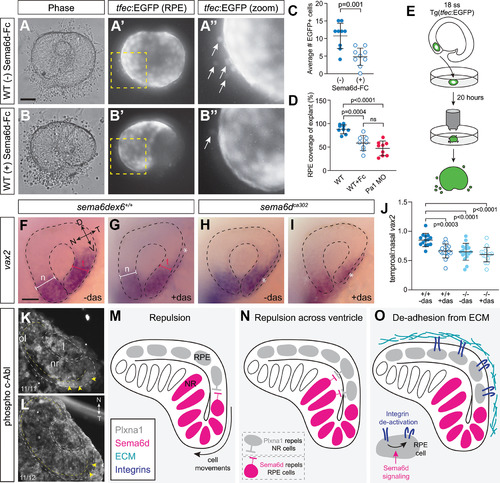
Disrupted RPE morphogenesis in EXPRESSION / LABELING:
PHENOTYPE:
|
|
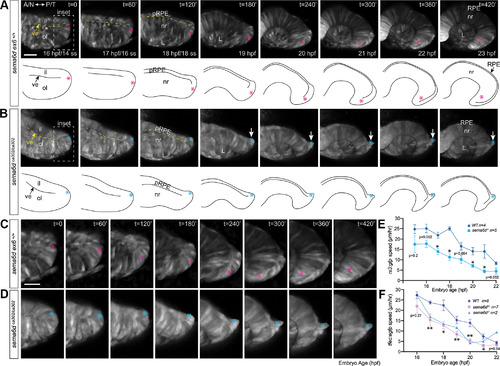
Temporal neural retina is disorganized in |
|
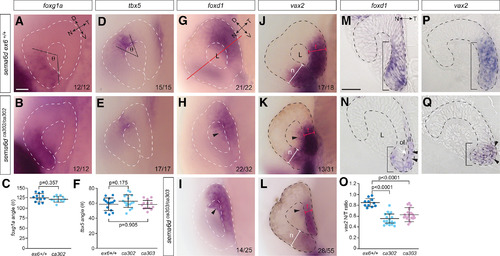
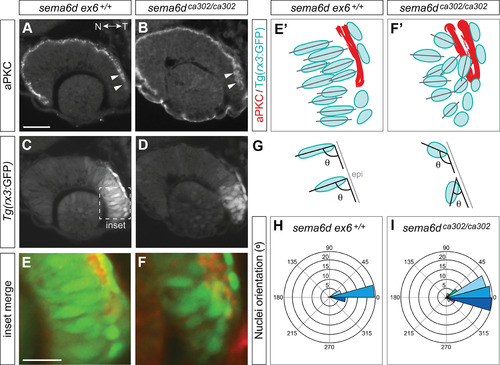
Ventral inner leaflet cells fail to move appropriately around the distal rim during optic cup morphogenesis. EXPRESSION / LABELING:
PHENOTYPE:
|
|
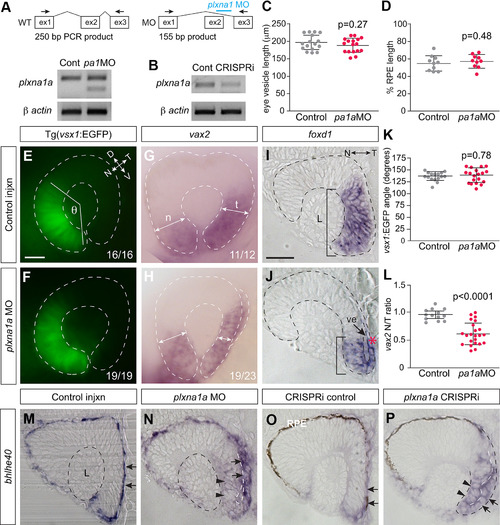
Plxna1a loss-of-function recapitulates the temporal eye defects observed in EXPRESSION / LABELING:
PHENOTYPE:
|
|
Possible Sema6d-Plxna1 repellent interactions during optic cup morphogenesis. |