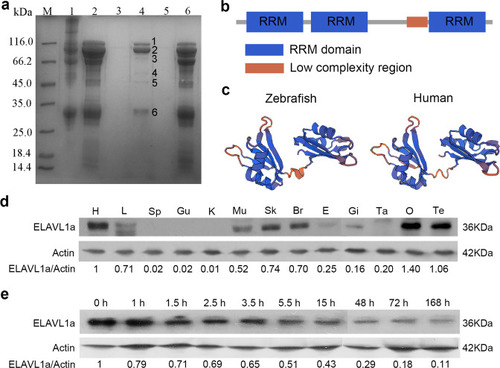
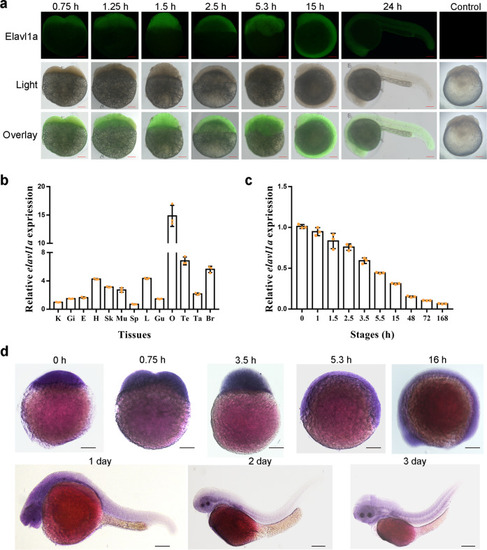
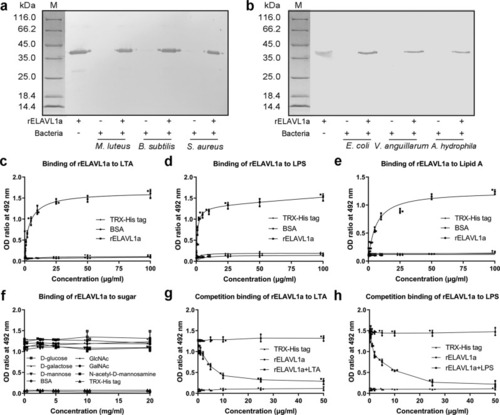
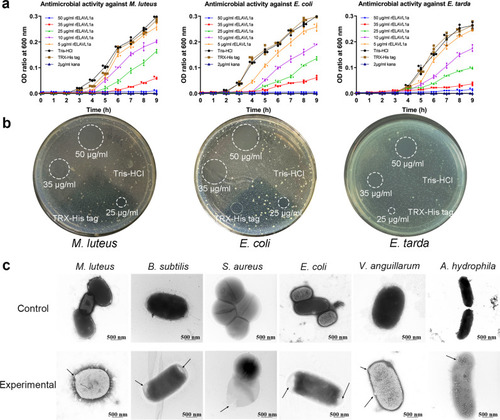
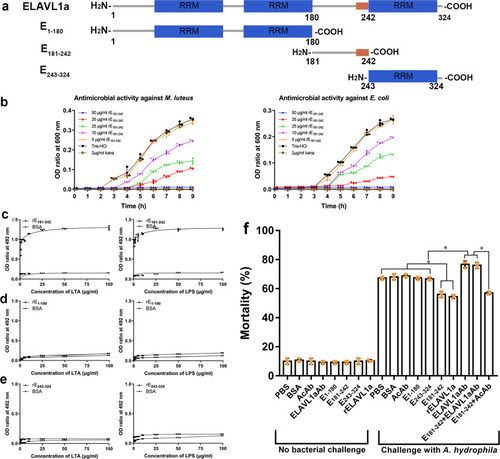
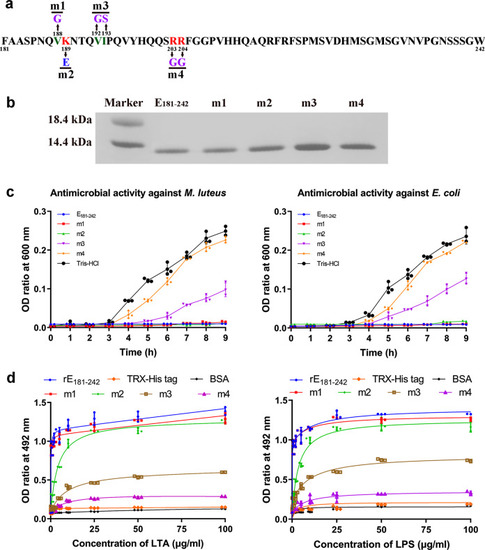
- Title
-
ELAVL1a is an immunocompetent protein that protects zebrafish embryos from bacterial infection
- Authors
- Ni, S., Zhou, Y., Song, L., Chen, Y., Wang, X., Du, X., Zhang, S.
- Source
- Full text @ Commun Biol
|
|
|
|
|
|
|
|
|
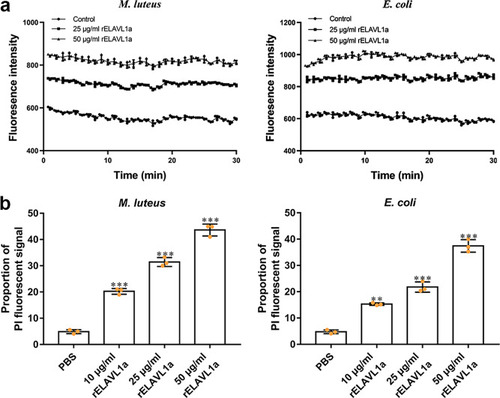
a The rELAVL1a caused depolarization of the bacterial plasma membrane. The changes in fluorescence intensity were recorded with a Tecan GENios plus spectrofluorometer at an excitation wavelength of 622 nm and an emission wavelength of 670 nm. Control, HEPES buffer containing 20 mM glucose. b The effects of rELAVL1a on the membrane integrity of M. luteus and E. coli, cells analyzed by flow cytometry. All data were expressed as mean values ± SD. (n = 3). The data are from three independent experiments performed in triplicate. The bars represent the mean ± S.D. The significance of the difference was determined by one-way ANOVA. **p < 0.01, ***p < 0.001. |
|
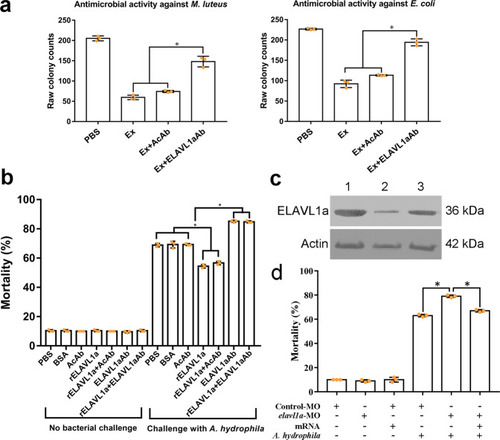
a Antimicrobial activities of the embryo extract against M. luteus and E. coli. b The early (8-cell stage) embryos were first microinjected with PBS, BSA, anti-β-actin antibody (AcAb), anti-ELAVL1a antibody (ELAVL1aAb), rELAVL1a, and rELAVL1a plus AcAb or rELAVL1a plus ELAVL1aAb and then challenged by injection with live A. hydrophila (8.3 × 107 cells/ml). The development of the embryos was observed, and the cumulative mortality rate was calculated at 24 h after bacterial injection. c Western blotting analysis of the efficacy of elavl1a-MO. β-Actin was used as control. Lane 1, 2, 3, the levels of ELAVL1a and β-Actin in the embryos injected with control morpholino (1), elavl1a-MO (2), or co-injection of elavl1a-MO plus elavl1a-mRNA (3). d The reduced antibacterial activity of the elavl1a-MO-knockdown embryos. The 24-h mortality of elavl1a-MO-knockdown group, control group, and rescued group, after the challenge with live A. hydrophila (8.3 × 107 cells/ml). All data were expressed as mean values ± S.D. (n = 3). The data are from three independent experiments performed in triplicate. The bars represent the mean ± SD. The significance of the difference was determined by one-way ANOVA. *p < 0.05. M, marker; con, control; Ex, embryo extract; AcAb, anti-β-actin antibody; ELAVL1aAb, mouse anti-ELAVL1a antibody. |
|
|
|
|