- Title
-
Identification of 3' UTR motifs required for mRNA localization to myelin sheaths in vivo
- Authors
- Yergert, K.M., Doll, C.A., O'Rouke, R., Hines, J.H., Appel, B.
- Source
- Full text @ PLoS Biol.
|
(A) Schematic representation of the MS2 mRNA reporter with motifs inserted upstream of the |
|
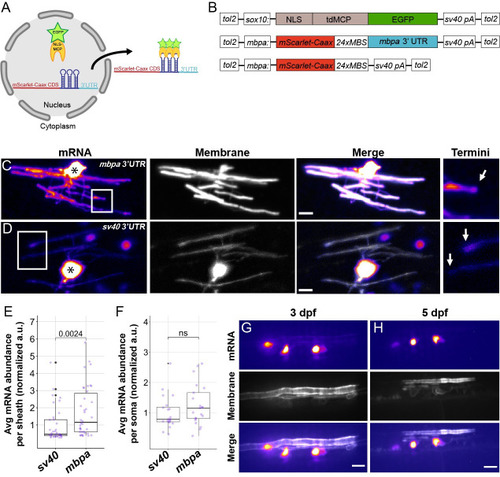
(A) Schematic of the MS2 system to visualize mRNA localization in oligodendrocytes. |
|
(A and B) Representative images of smFISH experiments using 4 dpf transgenic larva expressing EGFP-CAAX to mark oligodendrocytes. Images show sagittal sections of the hindbrain. DAPI stain labels nuclei. Sections were treated with smFISH probes designed to detect |
|
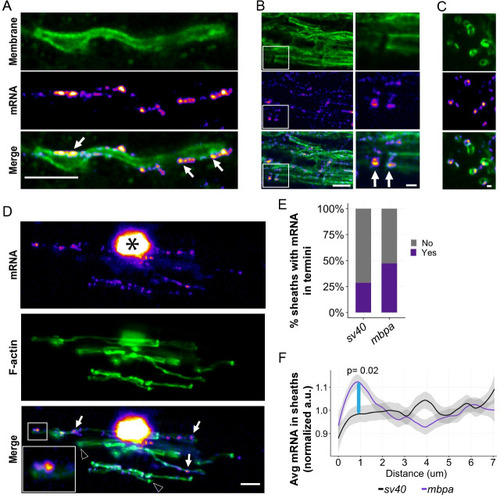
(A) smFISH images of a single optical section of a myelin sheath in a 3-dpf larva spinal cord. |
|
(A) Work flow to identify 3′ UTR candidates from RNA-seq data [ |
|
Representative images of smFISH experiments to visualize |
|
(A) Schematic representation of the 3 motifs identified in the 3′ UTRs of |
|
(A) Schematic representation of motifs 1, 2, or 3 enrichment in the myelin transcriptome in comparison to length-matched, randomized sequences using MEME suite Analysis of Motif Enrichment (version 5.1.1) [ |