|
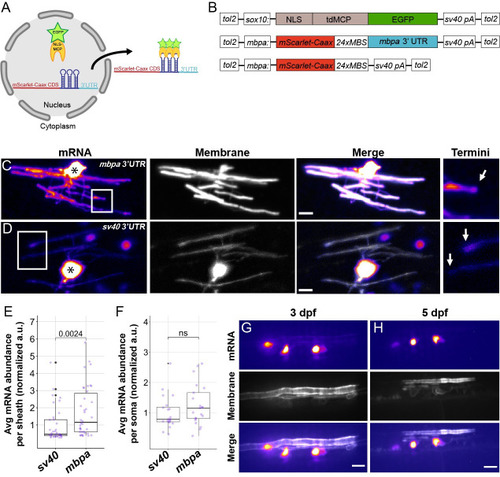
The <italic>mbpa</italic> 3′ UTR is sufficient to localize mRNA to myelin sheaths in living zebrafish.(A) Schematic of the MS2 system to visualize mRNA localization in oligodendrocytes. sox10 regulatory DNA drives expression of nuclear-localized MS2 coat protein, NLS-MCP-EGFP (orange crescent and green star). mbpa regulatory elements drive expression of mRNA encoding mScarlet-CAAX fluorescent protein with a repetitive sequence that creates 24 stem loops (24xMBS). When co-expressed, the mRNA–protein complex is exported from the nucleus and localized via the 3′ UTR. (B) Schematic of MS2 expression plasmids used for transient expression in oligodendrocytes with target sequences for Tol2 transposase to facilitate transgene integration. (C and D) Representative images of localization directed by the mbpa (C) or control sv40 3′ UTR (D). Asterisks mark cell bodies with high expression levels of the nuclear-localized MCP-EGFP. Boxed areas are enlarged to highlight sheath termini (arrows). (E) Average mRNA abundance per myelin sheath, measured by EGFP fluorescence intensity normalized to the average intensity of the sv40 control. sv40: n = 5 larvae, 35 sheaths. mbpa: n = 6 larvae, 38 sheaths. (F) Average mRNA abundance per soma, measured by EGFP fluorescence intensity normalized to the average intensity of the sv40 control. sv40: n = 11 larvae, 20 cell bodies. mbpa: n = 15 larvae, 21 cell bodies. (G and H) Representative images of 2 myelinating oligodendrocytes expressing mRNA lacking the 24xMBS. NLS-MCP-EGFP remains in the nucleus at 3 dpf (G) and 5 dpf (H). Scale bars, 10 μm. Statistical significance evaluated using Wilcoxon test. The underlying numerical data can be found in S1 and S2 Data. 3' UTR, 3' untranslated region; dpf, days post fertilization.
|