- Title
-
Sanguisorba officinalis L. Suppresses Triple-Negative Breast Cancer Metastasis by Inhibiting Late-Phase Autophagy via Hif-1α/Caveolin-1 Signaling
- Authors
- Wang, N., Muhetaer, G., Zhang, X., Yang, B., Wang, C., Zhang, Y., Wang, X., Zhang, J., Wang, S., Zheng, Y., Zhang, F., Wang, Z.
- Source
- Full text @ Front Pharmacol
|
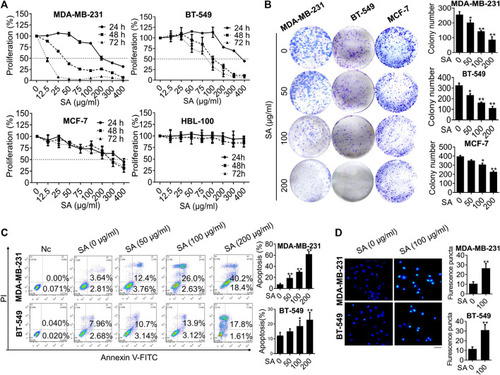
SA exerts anti-proliferation and apoptosis-promoting effects in breast cancer cells. |
|
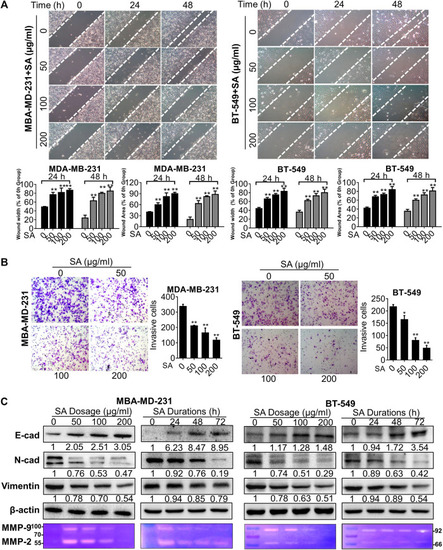
SA suppresses migration and invasion in metastatic cells. |
|
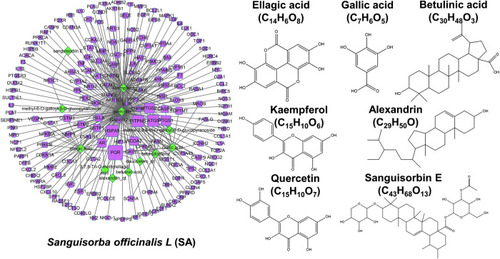
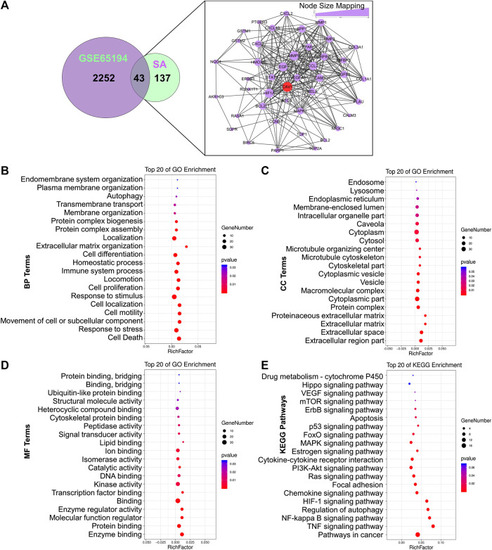
Network pharmacology analysis of SA. |
|
Establishment of an ingredient-target-breast cancer network of SA. |
|
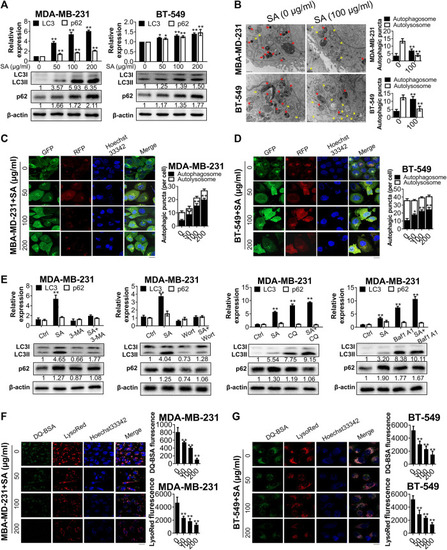
SA inhibits autophagic flux in metastatic cells. |
|
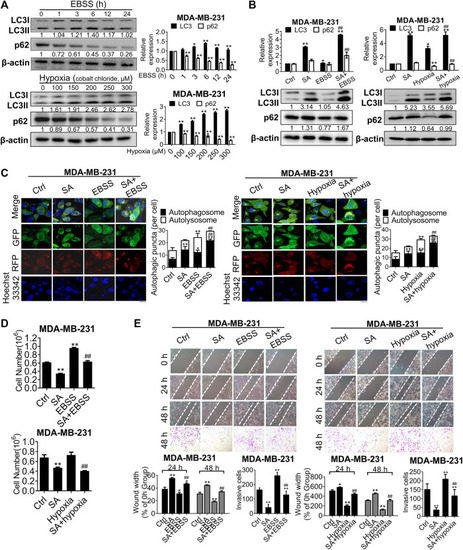
SA suppresses autophagy-mediated metastatic processes during starvation or hypoxia. |
|
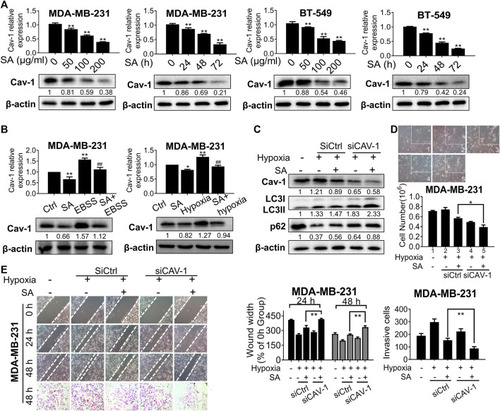
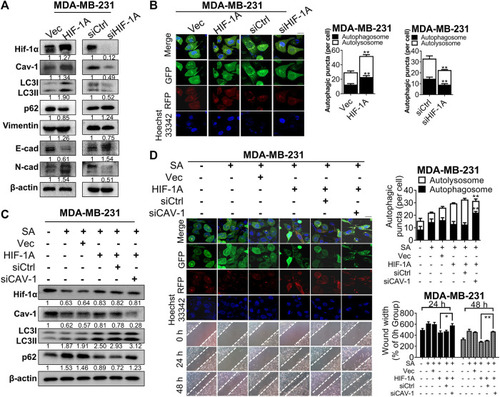
SA inhibits Cav-1 expression and blocks late-phase autophagy and subsequent metastatic processes. |
|
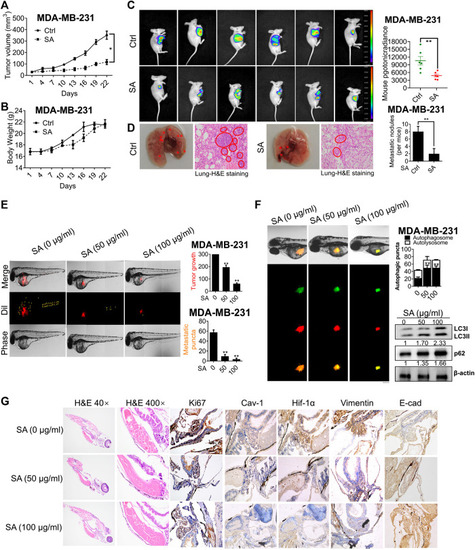
SA exerts anti-metastatic effects partially |
|
SA suppresses breast cancer growth and metastasis |