- Title
-
µSPIM toolset: A software platform for selective plane illumination microscopy
- Authors
- Saska, D., Pichler, P., Qian, C., Buckley, C.L., Lagnado, L.
- Source
- Full text @ J. Neurosci. Methods
|
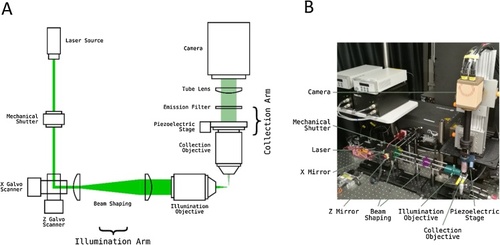
Light-Sheet Microscope Implementation. |
|
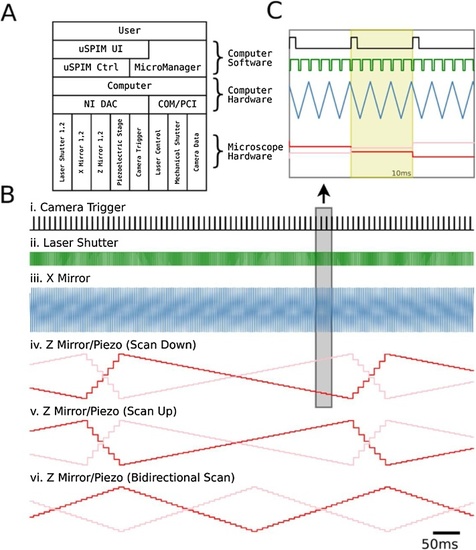
Light-Sheet Microscope Hardware Control: (A) Interaction of different components of the μSPIM Toolset-based setup in a typical acquisition setup. μSPIM Toolset provides control and synchronization of hardware through NI DAC with MicroManager controlling the camera and mechanical shutters through PCI and COM ports. |
|
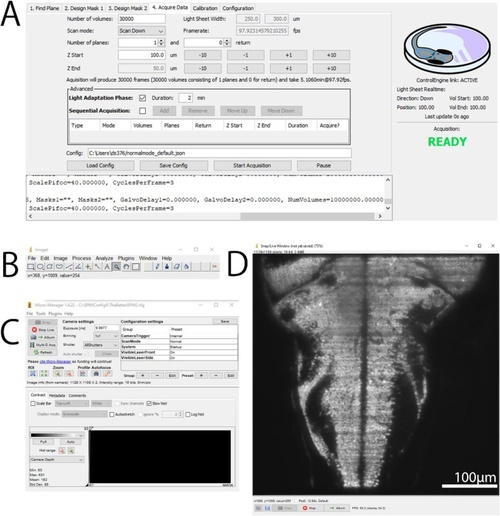
μSPIM Toolset User Interface. Following a common MicroManager design, the control interface is separated into several components: μSPIM Toolset-provided plugin window with control over the light-sheet generation including calibration and acquisition routines shown in A, basic ImageJ tools shown in B and MicroManager interface providing control over the acquisition hardware shown in C and live view of the camera shown in D. The shown user interface has been captured during the acquisition from a 7 dpf larval zebrafish from the |
|
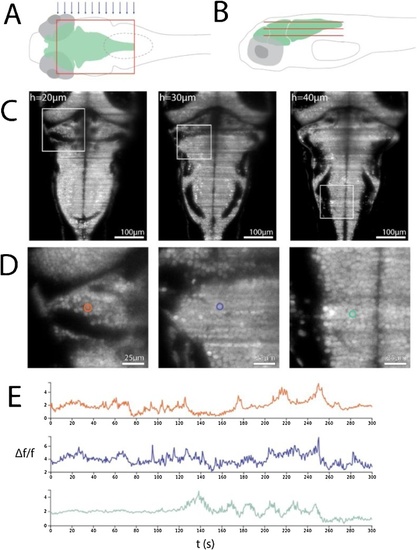
Imaging larval zebrafish: (A and B). Schematic of a single light sheet covering the hindbrain of a larval zebrafish from above (A) as well as a number of light sheets (constituting a volume) from the side (B, sheet size and spacing are not to scale). |
|
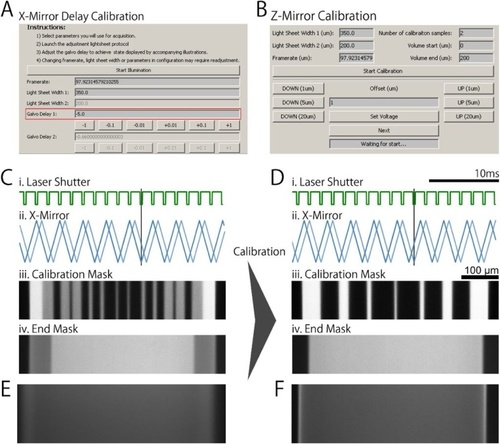
μSPIM Toolset Calibration μSPIM Toolset provides the user with calibration tools which can be used to assess and correct the performance of the individual microscope elements. |
|
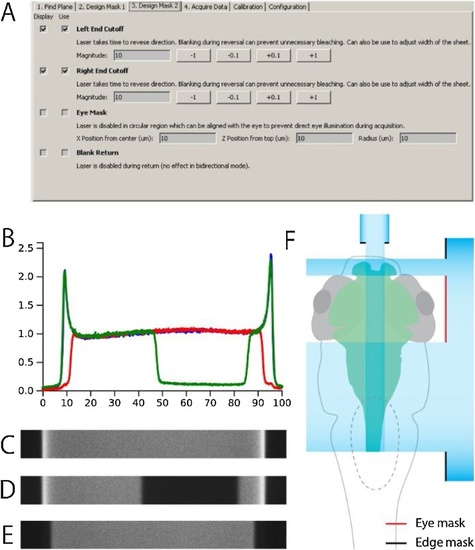
Light-Sheet Laser Masks. |
|
Acquisition using μSPIM Toolset & Output Data Format. μSPIM Toolset generates a separate metadata file (shown in |