- Title
-
Genetic Deletion of miR-430 Disrupts Maternal-Zygotic Transition and Embryonic Body Plan
- Authors
- Liu, Y., Zhu, Z., Ho, I.H.T., Shi, Y., Li, J., Wang, X., Chan, M.T.V., Cheng, C.H.K.
- Source
- Full text @ Front Genet
|
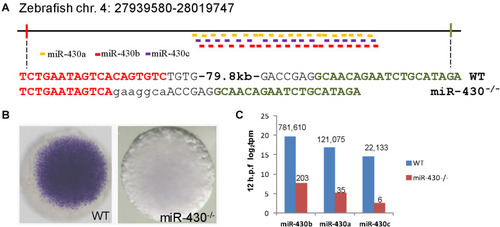
Targeted deletion of |
|
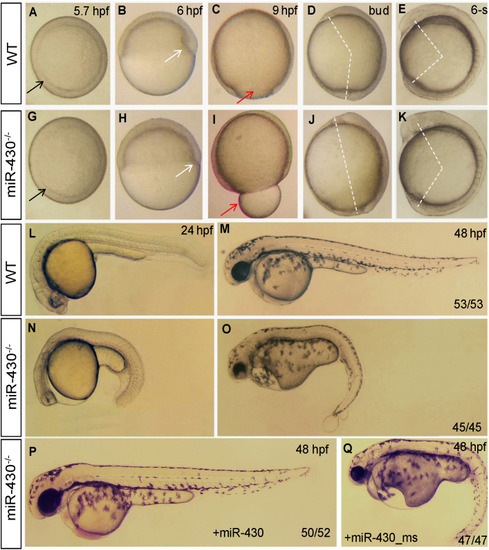
Morphological defects in the |
|
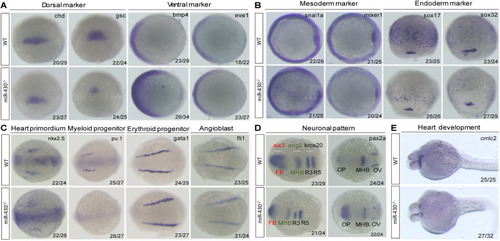
Marker gene analysis. Marker gene expression of the WT and |
|
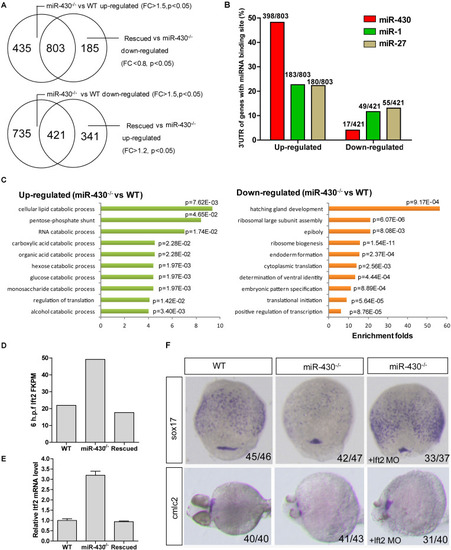
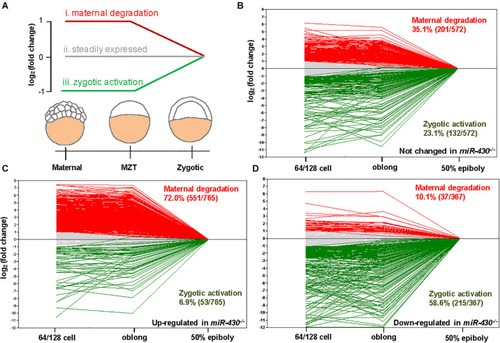
Transcriptome analysis. Total mRNAs of embryos from WT, |
|
|
|
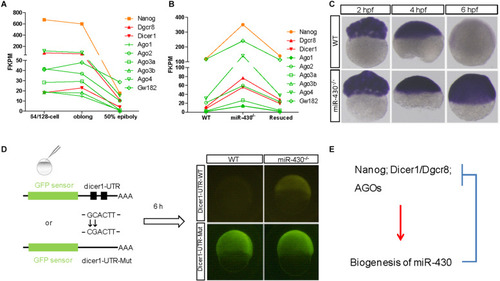
A reciprocal regulatory loop for |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

Unillustrated author statements PHENOTYPE:
|