- Title
-
Srag regulates autophagy via integrating into a preexisting autophagy pathway in testis
- Authors
- Cheng, Y., Lai, F., Wang, X., Shang, D., Zou, J., Luo, M., Xia, X., Cheng, H., Zhou, R.
- Source
- Full text @ Mol Bio Evol
|
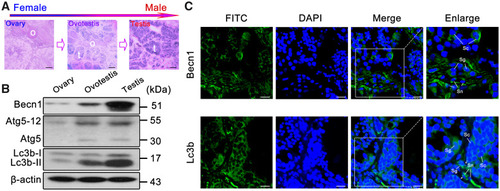
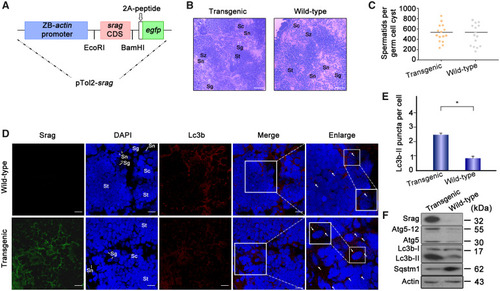
Upregulation of autophagy in testis. ( |
|
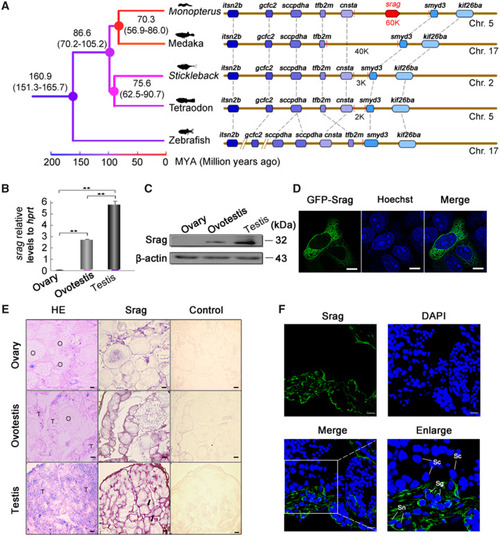
Identification and expression of |
|
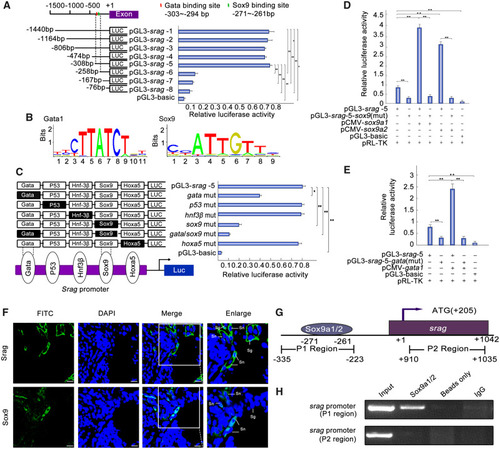
Sox9a1/2 and Gata1 upregulates |
|
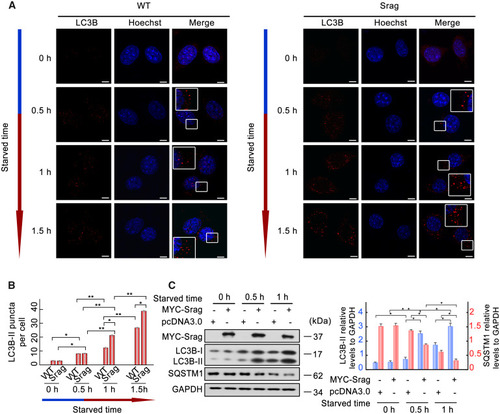
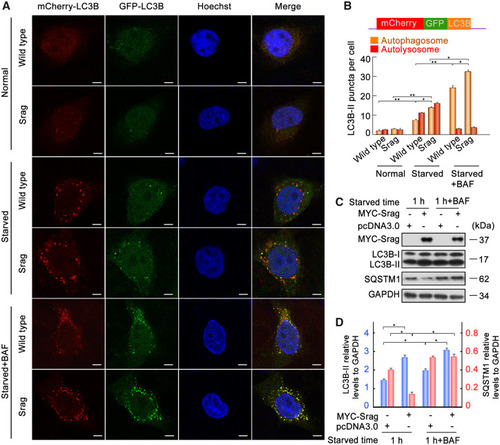
Srag overexpression promotes autophagy upon starvation induction. ( |
|
|
|
Srag-associated autophagy flux. ( |
|
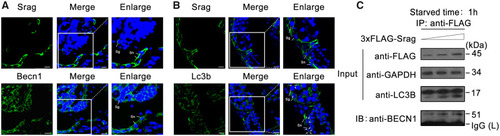
Srag coexpression with Becn1 and Lc3b and interaction between Srag and Becn1. ( |
|
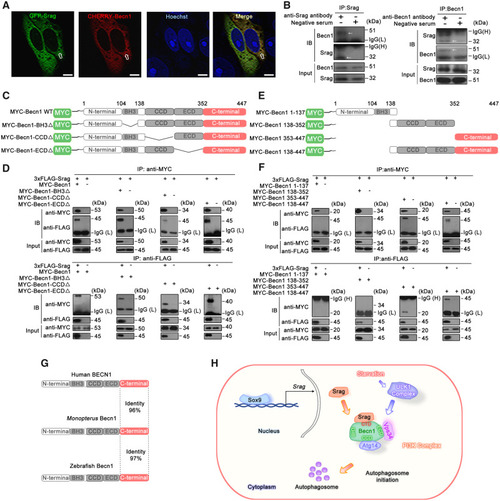
Srag interacts with Becn1. ( |