- Title
-
Hierarchical Compression Reveals Sub-Second to Day-Long Structure in Larval Zebrafish Behavior
- Authors
- Ghosh, M., Rihel, J.
- Source
- Full text @ eNeuro
|
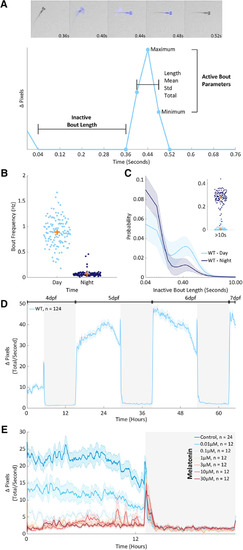
Behavior at scale. |
|
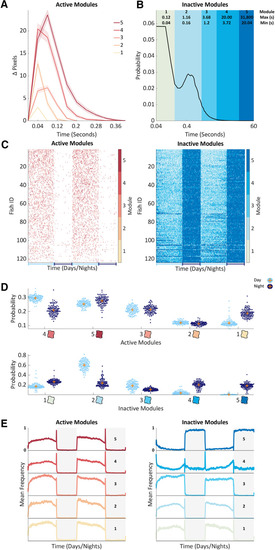
Unsupervised learning identifies contextual behavioral modules. |
|
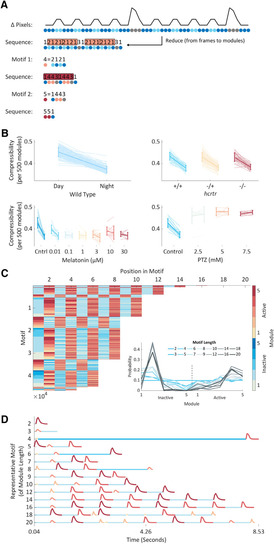
Hierarchical compression reveals structure in zebrafish behavior. |
|
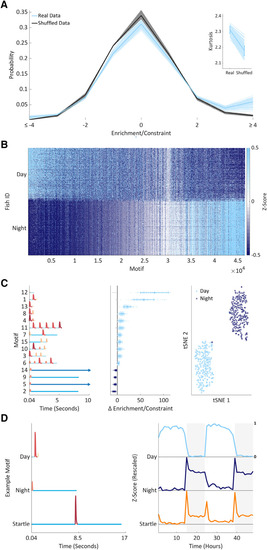
Supervised learning identifies contextual behavioral motifs. |
|
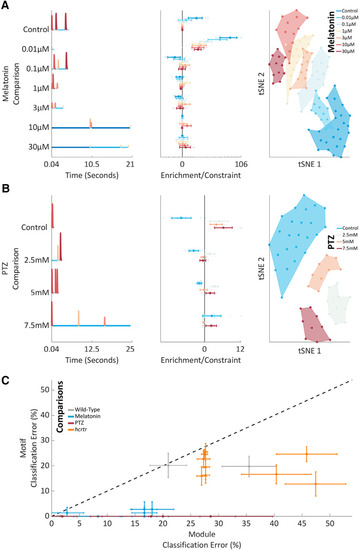
Pharmacological behavioral motifs. |