- Title
-
Structural Neural Connectivity Analysis in Zebrafish With Restricted Anterograde Transneuronal Viral Labeling and Quantitative Brain Mapping
- Authors
- Ma, M., Kler, S., Pan, Y.A.
- Source
- Full text @ Front. Neural Circuits

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
Examples of third-order neuron labeling. Red arrowheads indicate third-order neurons labeled in the hindbrain by TRAS. Yellow dash line marks the border of mesencephalon and cerebellum. Scale bar: 50 μm. |
|
Retina injection and tissue clearing. (A)Examples of RFP expression in the retina of unpigmented (PTU-treated) animals 3 days after virus injection into the temporal retina. Images are lateral views, with rostral side to the left and dorsal side to the top. Yellow dashed lines outline the eye (outer oval) and lens (inner oval). (B,C) Tissue clearing with sRIMS solution. Whole-mount ERK1/2 immunolabeling without (B) or with sRIMS clearing (C). Orthogonal views (XY, XZ, and YZ) of confocal image stacks are shown, centered just bellowed the cerebellum (intersect of yellow lines). Ventral structures are not visible without sRIMS clearing. Scale bars are 100 μm. |
|
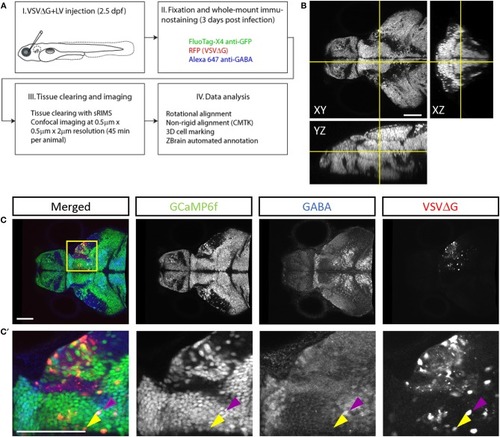
TRAS labeling of GCaMP6f positive and negative cells. A single confocal imaging plane is shown, with merged, GCaMP6f, GABA, and VSVΔG channels as indicated. Boxed area in (A) is shown in higher magnification in (A'). The purple arrowheads mark two GCaMP6f+/GABA+/VSVΔG+ inhibitory neurons. The orange arrowhead marks a GCaMP6f–/GABA–/VSVΔG+ cell. Scale bars are 100 μm. |
|
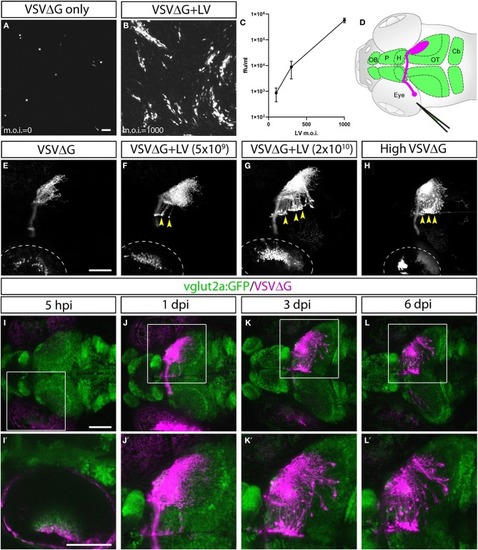
Lentivirus enabled |
|
Efferent projections of retinorecipient cells. |
|
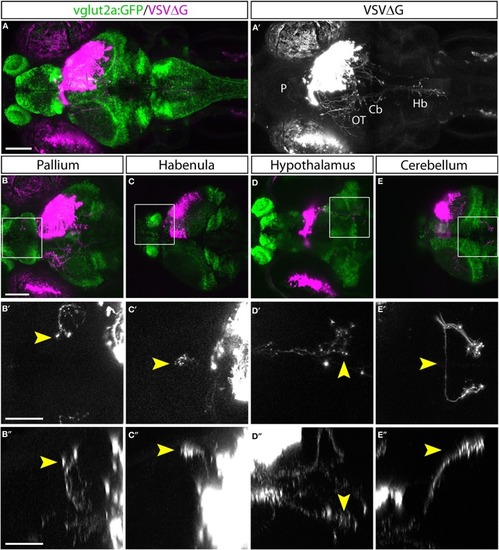
Cell-type characterization of TRAS labeling. |
|
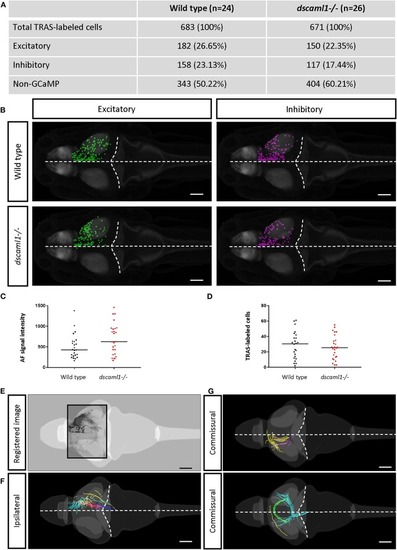
Annotation of TRAS-labeled neurons and efferent axons in the Z-Brain standard brain reference. |
|
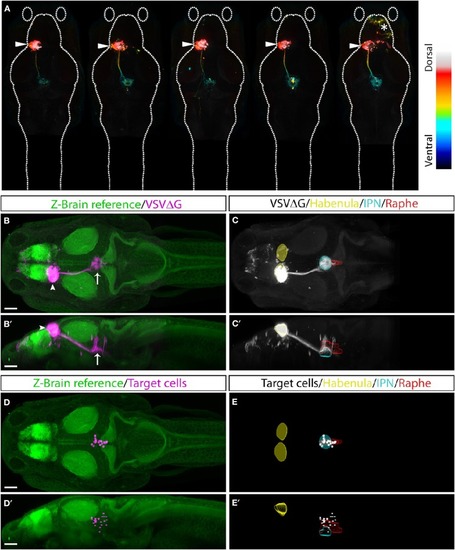
TRAS labeling of habenular target cells. |