- Title
-
Functional architecture underlying binocular coordination of eye position and velocity in the larval zebrafish hindbrain
- Authors
- Brysch, C., Leyden, C., Arrenberg, A.B.
- Source
- Full text @ BMC Biol.
|
Setup and circuit overview. |
|
Experimental strategy to assess binocular coordination. |
|
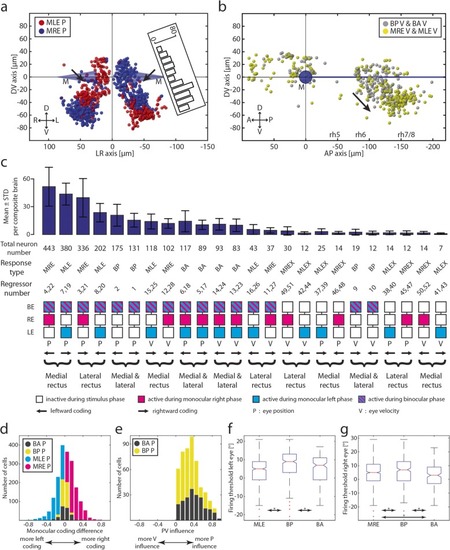
Monocular and binocular cell maps. |
|
Monocular/binocular synopsis. |
|
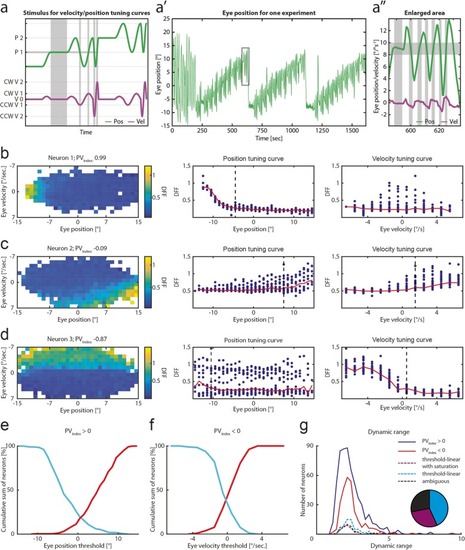
Neuronal tuning for eye velocity and position. |
|
PVIndex distribution and spatial location of identified neurons. |
|
Summary for binocular coordination and PV encoding in the larval zebrafish hindbrain. |