- Title
-
Endothelin signalling mediates experience-dependent myelination in the CNS
- Authors
- Swire, M., Kotelevtsev, Y., Webb, D.J., Lyons, D.A., Ffrench-Constant, C.
- Source
- Full text @ Elife
|
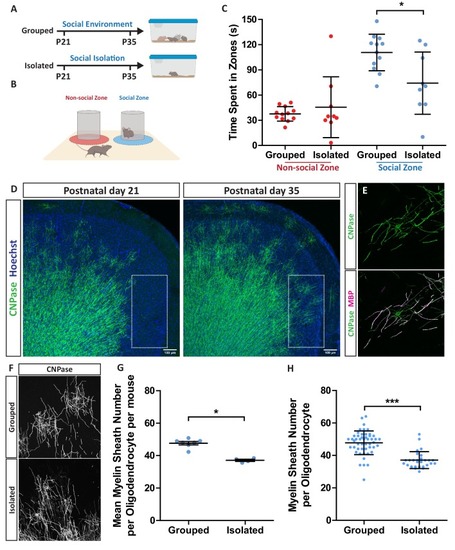
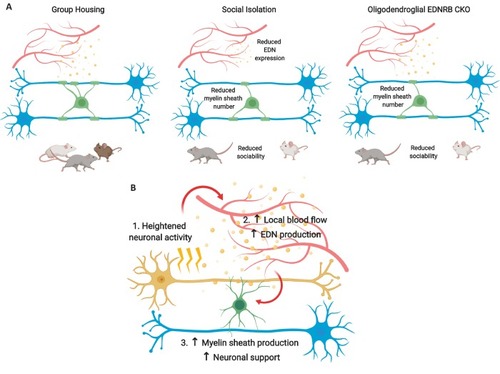
(A) Timeline for social isolation experiment. At postnatal day 21 male mice were housed in a social environment containing 3–5 mice or on their own in isolation. Mice were analysed at P35. (B) Schematic of social interaction assay. Mice were recorded for 5 min exploring an arena containing two identical wire mesh containers: one container housed an unrelated male wild type mouse (social zone), while the other remained empty (non-social zone). (C) Time spent within 2.5 cm of non-social container: Grouped 37.58 s ± 8.683 n = 12, Isolated 45.59 ± 36.28 n = 9 and social container: Grouped 110.7 s ± 21.71 n = 12, Isolated 74.24 s ± 37.07 n = 9 (mean ± standard deviation). Unpaired T-test p=0.0107. (D) Coronal section of mouse prefrontal cortex stained for CNPase and nuclei. Layers II/III of the medial prefrontal cortex outlined by dashed box. Scale bars = 100 μm. (E) Layer II/III oligodendrocyte stained for CNPase and MBP. (F) Representative images of medial prefrontal cortex oligodendrocytes stained for CNPase. (G) Mean number of myelin sheaths formed by oligodendrocytes per mouse. Grouped 47.66 ± 1.015 n = 7 mice, Isolated 37.11 ± 0.6425 n = 4 mice (mean ± standard error). Mann-Whitney test, p=0.0106. (H) Pooled data for number of myelin sheaths formed by layer II/III medial prefrontal cortex oligodendrocytes. Grouped 47.80 ± 7.289 n = 49 cells from seven mice, Isolated 37.11 ± 5.202 n = 28 cells from four mice (mean ± standard deviation). Mann-Whitney test, p=<0.001 |
|
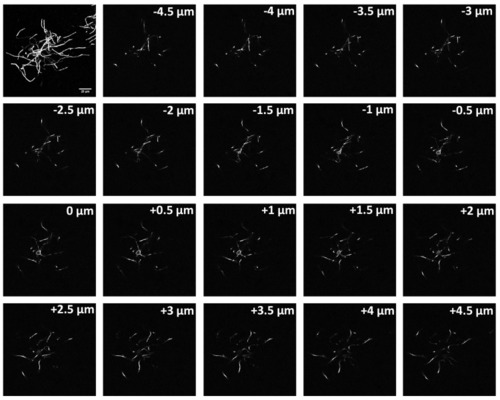
A full maximum intensity projection of a complete a layer II/III oligodendrocytes in mouse cortex stained for CNPase followed by 0.5 µm slices. Tracing the fine processes through each slice enables assessment of the myelin sheaths formed by individual oligodendrocytes. |
|
( |
|
( |
|
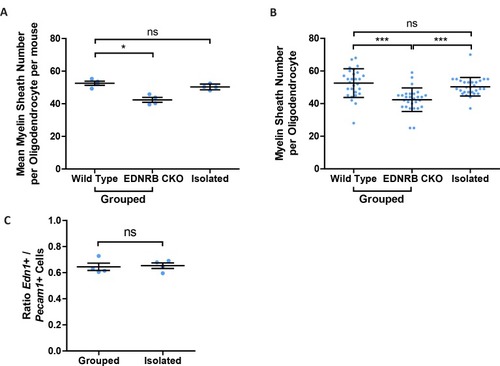
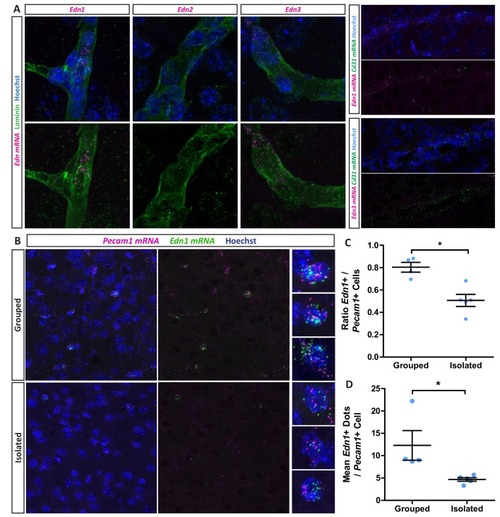
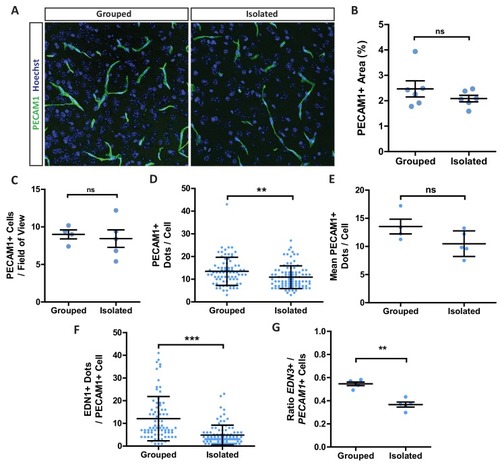
(A) Edn1 and Edn3 mRNA expression in laminin positive and CD31 positive blood vessels as revealed by RNAScope in situ hybridisation. (B) Representative images of Edn1 and Pecam1 mRNA expression in the mPFC. (C) Quantification of the number of Edn1 expressing Pecam1 positive endothelial cells. Grouped 0.8033 ± 0.04411 n = 4 mice, Isolated 0.5074 ± 0.05412 n = 5 mice (mean ± standard error). Mann-Whitney test, p=0.0159. (D) Quantification of the mean Edn1 mRNA molecules expressed by Pecam1 positive cells per mouse. Grouped 12.29 ± 3.312 n = 4 mice, Isolated 4.673 ± 0.4059 n = 5 mice (mean ± standard error). Mann-Whitney test, p=0.0159. |
|
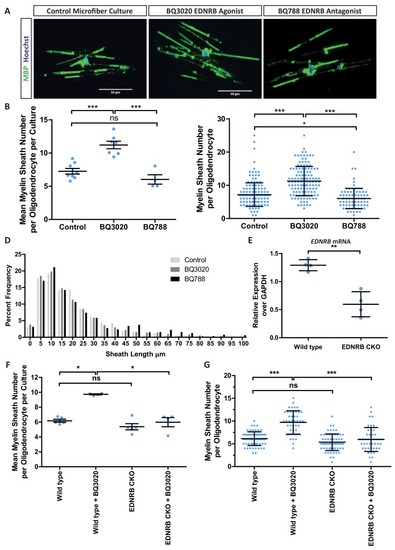
Expression of |
|
( |
|
( |
|
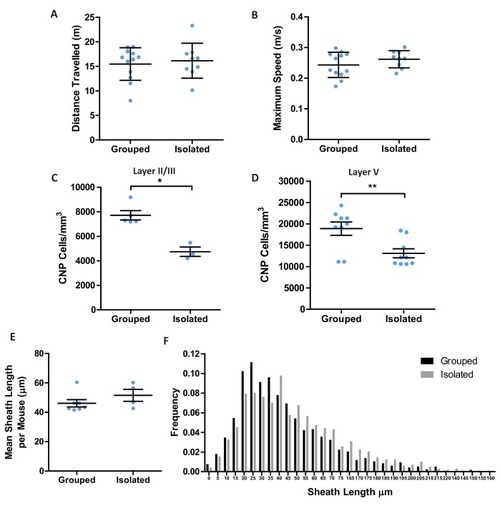
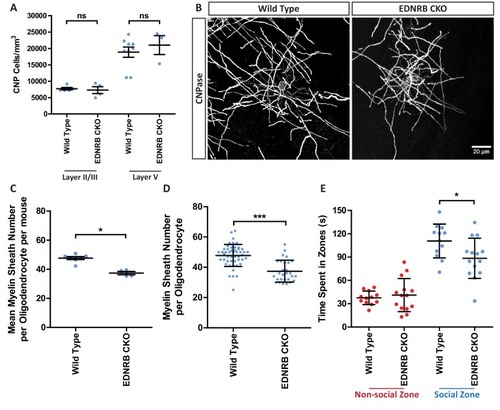
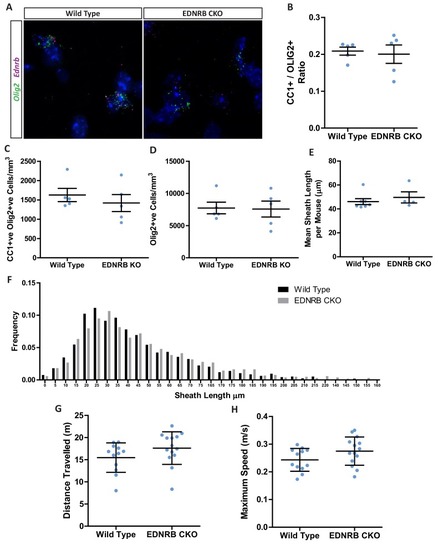
(A) Quantification of CNP positive cells in medial prefrontal cortex layers II/III: Wild type 7709 ± 378.7 n = 5 mice, EDNRB CKO 7288 ± 1054 n = 4 mice (mean ± standard error) and layer V: Wild type 18879 ± 1559 n = 9 mice, EDNRB CKO 21016 ± 2878 n = 3 mice (mean ± standard error). Mann Whitney test, layer II/III p=0.9048, layer V p=0.3527. (B) Representative images of medial prefrontal cortex oligodendrocytes stained for CNPase. Scale bar = 20 μm. (C) Mean number of myelin sheath formed by oligodendrocytes per mouse. Wild type 47.66 ± 1.015 n = 7 mice, EDNRB CKO 37.39 ± 1.099 n = 4 mice (mean ± standard error). Mann-Whitney test, p=0.0106. (D) Pooled data for number of myelin sheaths formed by layer II/III medial prefrontal cortex oligodendrocytes. Wild type 47.80 ± 7.289 n = 49 cells from seven mice, EDNRB CKO 37.39 ± 7.208 n = 28 cells from four mice (mean ± standard deviation). Mann-Whitney test, p=<0.001. (E) Time spent within 2.5 cm of non-social container: Wild type 37.58 s ± 8.683 n = 12, EDNRB CKO 41.06 ± 21.29 n = 14 and social container: Wild type 110.7 s ± 21.71 n = 12, EDNRB CKO 88.39 s ± 25.79 n = 14 (mean ± standard deviation). Unpaired T-test p=0.0267 |
|
( |
|
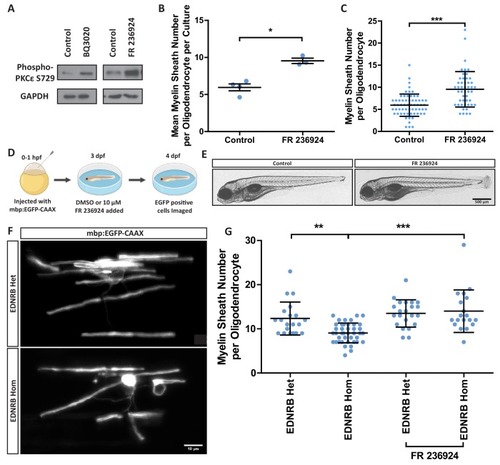
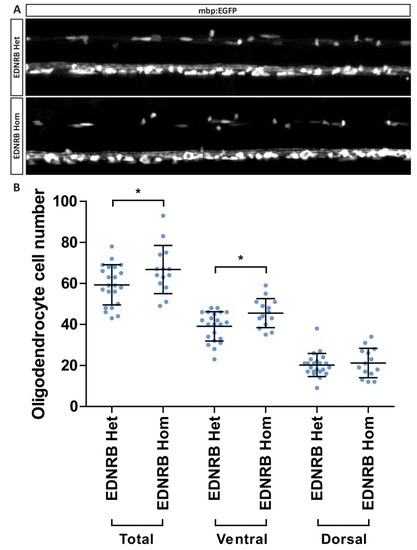
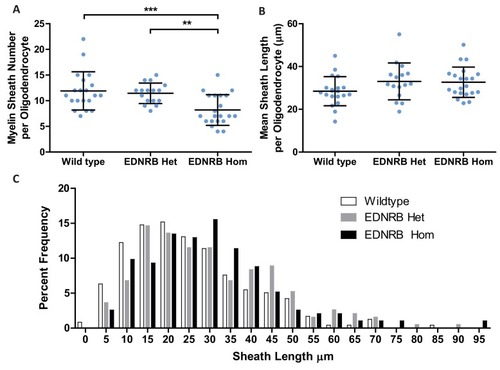
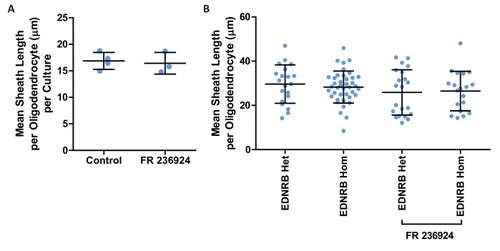
(A) Western blot images of rat oligodendrocytes treated with EDNRB agonist BQ3020 and PKCε agonist FR 236924 for 15 min. Antibodies used: Phosphorylated PKCε S729 and loading control GAPDH. (B) Mean number of myelin sheaths formed by rat oligodendrocytes on microfibers per experiment. Control 5.959 ± 0.4708 n = 4, FR 236924 9.542 ± 0.3614 n = 3 (mean ± standard error). Unpaired T-test p=0.0024. (C) Pooled data for number of myelin sheaths formed by rat oligodendrocytes on microfibers. Control 5.952 ± 2.525 n = 62 cells from four experiments, FR 236924 9.542 ± 4.016 n = 48 from three experiments (mean ± standard deviation). Mann-Whitney test, p=<0.001. (D) Schematic for zebrafish larvae treatment with FR 236924. (E) Representative images of 4 dpf zebrafish larvae treated with DMSO control or FR 236924. Scale bar = 500 μm. (F) Representative images of mbp:EGFP-CAAX oligodendrocytes in four dpf zebrafish larvae. Scale bar = 10 μm. (G) Pooled data for number of myelin sheaths formed by zebrafish oligodendrocytes. EDNRB Het 12.35 ± 3.746 n = 20 cells, EDNRB Hom (rse) 9.073 ± 2.229 n = 41 from four experiments, EDNRB Het + FR 236924 13.5 ± 3.098 n = 22 cells from five experiments, EDNRB Hom + FR 236924 14 ± 4.807 n = 19 cells (mean ± standard deviation). 1-way ANOVA PHENOTYPE:
|
|
( PHENOTYPE:
|
|
( PHENOTYPE:
|
|
( |
|
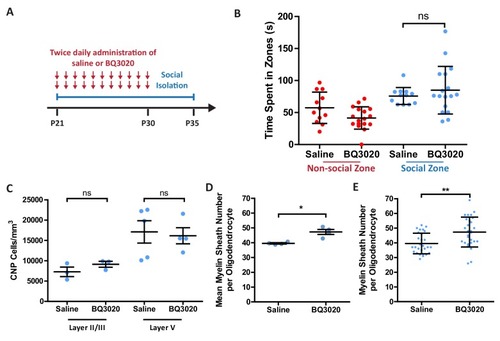
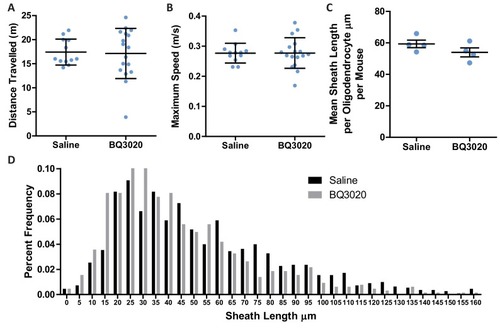
(A) Timeline for the intranasal experiment. At postnatal day 21 male mice were housed on their own in isolation. Mice were given two daily administrations of saline or EDNRB agonist BQ3020 from P21-P30. Mice were analysed at P35. (B) Time spent within 2.5 cm of non-social container: Saline 57.42 s ± 24.44 n = 12, BQ3020 41.44 ± 17.43 n = 17 and social container: Saline 75.82 s ± 13.26 n = 12, BQ3020 84.96 s ± 37.20 n = 17 (mean ± standard deviation). T-test. (C) Quantification of CNP positive cells in medial prefrontal cortex layers II/III: Saline 7278 ± 1165 n = 3 mice, BQ3020 9142 ± 713.3 n = 3 mice (mean ± standard error) and layer V: Saline 17114 ± 2750 n = 5 mice, BQ3020 16154 ± 1980 n = 4 mice (mean ± standard error). (D) Mean number of myelin sheath formed by oligodendrocytes per mouse. Saline 39.54 ±0.4301 n = 4 mice, BQ3020 47.29 ± 1.687 n = 4 mice (mean ± standard error). Mann-Whitney test, p=0.0286. (E) Pooled data for number of myelin sheaths formed by layer II/III medial prefrontal cortex oligodendrocytes. Saline 39.54 ±6.973 n = 28 cells from four mice, BQ3020 47.29 ± 10.18 n = 28 cells from seven mice (mean ± standard deviation). Mann-Whitney test, p=0.0019. |
|
( |
|
( |