- Title
-
Screening for insulin-independent pathways that modulate glucose homeostasis identifies androgen receptor antagonists
- Authors
- Mullapudi, S.T., Helker, C.S., Boezio, G.L., Maischein, H.M., Sokol, A.M., Guenther, S., Matsuda, H., Kubicek, S., Graumann, J., Yang, Y.H.C., Stainier, D.Y.
- Source
- Full text @ Elife

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
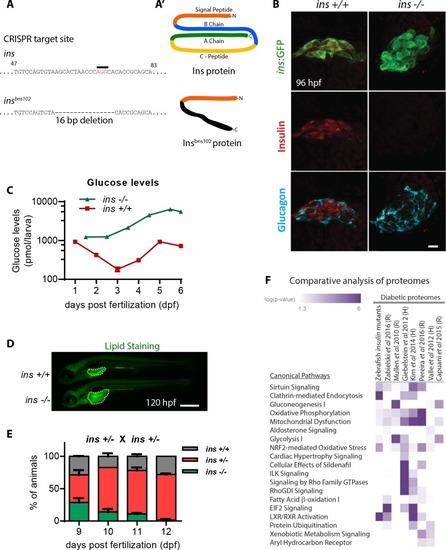
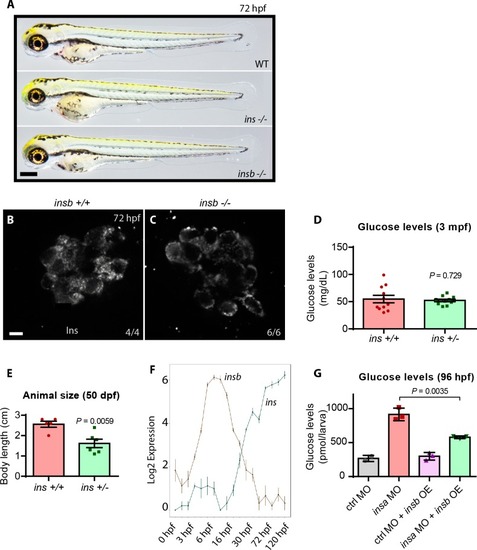
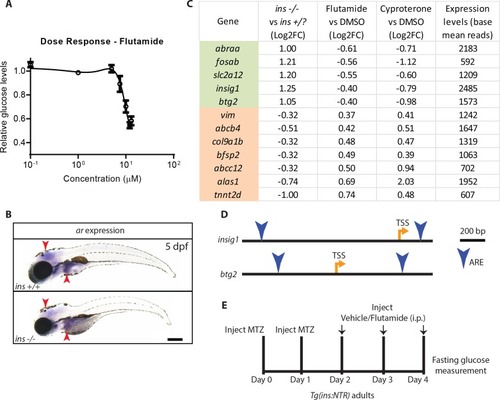
(A) CRISPR target site in the insulin gene, with PAM sequence highlighted, and the resulting 16 bp deletion allele (below). (A’) Schematic of wild-type Insulin protein and the predicted mutant protein which contains novel sequence (black). (B) Confocal projection images of the pancreatic islet in 96 hpf Tg(ins:GFP) ins +/+ and ins -/- animals immunostained for Insulin (red) and Glucagon (cyan). (C) Free glucose levels in wild-type and mutant animals from 1 to 6 dpf; mean ± SEM, n = 2–4 replicates. (D) Nile Red staining (green) for neutral lipids in 120 hpf wild-type (top) and mutant (bottom) larvae, with yolk lipid content outlined (yellow dots). (E) Genotype distribution from ins ± incross, calculated as the percentage of total animals at each stage; mean ± SEM, n = 32 animals at each stage. (F) Heat map of the proteomic signature of zebrafish ins mutants at 120 hpf compared to signatures from rodent (R) and human (H) diabetic proteome studies. Canonical pathways implicated in most studies are listed first. P-value cut-off set at <0.05. Scale bars: 10 μm (B), 500 μm (D). |
|
( PHENOTYPE:
|
|
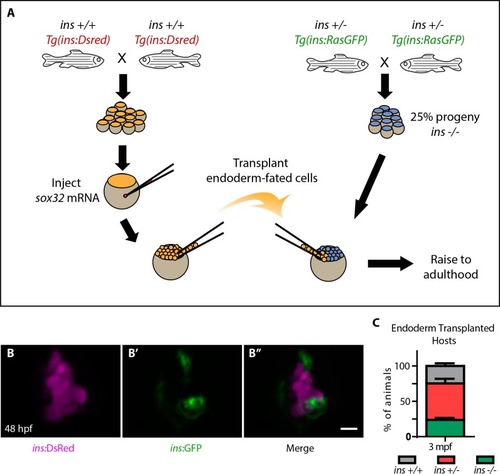
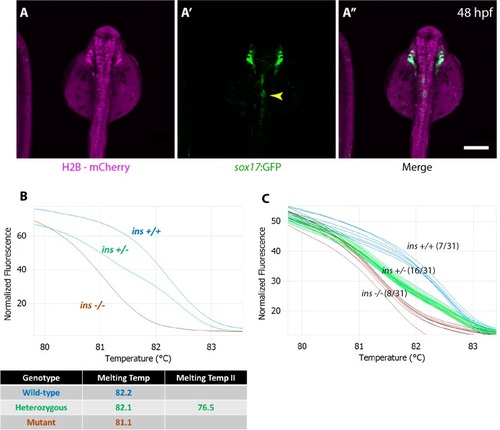
(A) Schematic depicting the endoderm transplantation protocol; sox32 mRNA-injected ins +/+ donor cells (orange) were transplanted into host embryos (blue) to form chimeric animals. (B–B’’) Confocal projection images of the pancreatic islet of a 48 hpf chimeric animal showing β-cells from the host (green, (B’) and the transplanted ins +/+ cells (magenta, (B). (C) Genotype distribution in the raised three mpf chimeric animals, determined by genotyping fin tissue; mean ± SEM, n = 3 transplant experiments, 18–32 animals per experiment. Scale bar: 10 μm. |
|
( |
|
( EXPRESSION / LABELING:
|