- Title
-
Functional analysis of the zebrafish ortholog of HMGCS1 reveals independent functions for cholesterol and isoprenoids in craniofacial development
- Authors
- Quintana, A.M., Hernandez, J.A., Gonzalez, C.G.
- Source
- Full text @ PLoS One
|
Mutation of hmgcs1 causes craniofacial abnormalities. Alcian staining was performed to visualize the developing cartilage in hmgcs1 wildtype or heterozygous (Sibling) or homozygous hmgcs1 mutant larvae (Mutant). Embryos were stained at 4 days post fertilization and manual dissection of the viscerocranium and neurocranium was performed. Viscerocranium is depicted in (A) and (B) and neurocranium is depicted in (A') and (B'). Asterisk denotes basihyal. n = 38 including 8 wildtype embryos, 6 homozygous mutants, and 24 heterozygous mutants. No significant defects were present in heterozygous carriers (S1 Fig). |
|
hmgcs1 does not regulate neural crest cell specification or migration. (A&B) In situ hybridization to detect sox10 expression was performed in hmgcs1 homozygous mutants or siblings at the 18 somite stage. Dorsal view with anterior to the left. (n = 15 including 8 wildtype/heterozygous siblings and 7 homozygous mutants). Wildtype Sibling is depicted in A, but no significant differences were observed between wildtype and heterozygous carriers. Arrowheads indicated two positive streams of Sox10+ positive cells. (C&D) Sagittal view of wildtype hmgcs1 siblings (hmgcs1 Sibling) or hmgcs1 homozygous mutant larvae (hmgcs1 MT). Larvae were maintained in a Tg(sox10:tagRFP) background and RFP was visualized using traditional microscopy at 1 day post fertilization (dpf) between Prim-5 and Prim-15. n = 20 including 15 wildtype and heterozygous individuals and 5 homozygous mutant individuals. Sibling depicted is a wildtype individual. No significant difference between wildtype embryos was detected. EXPRESSION / LABELING:
PHENOTYPE:
|
|
hmgcs1 modulates cranial neural crest cell differentiation. (A&B) Whole mount in situ hybridization (ISH) was performed at 1 day post fertilization (dpf) between Prim-5 and Prim- 15 on wildtype/heterozyous hmgcs1 siblings (Sibling) or hmgcs1 homozygous mutant larvae (Mutant) using a riboprobe specific to dlx2a. n = 20 including 16/20 wildtype and heterozygous siblings and 4/20 homozygous mutants. Panel A depicts a wildtype sibling and no significant difference between wildtype embryos was detected. (C&D) ISH was performed at 3 dpf (protruding mouth stage) on hmgcs1 siblings (wildtype and heterozygous individuals) or hmgcs1 homozygous mutant larvae with a riboprobe specifically detecting col2a1a expression. Numbers denote the pharyngeal arch patterns present a 3dpf. Arrowhead denotes a loss of defined expression of col2a1a in hmgcs1 mutant larvae. n = 36 including 29 wildtype and heterozygous individuals with 7 homozygous mutants. Panel C depicts a wildtype sibling with no significant differences observed between wildtype and heterozygous carriers. (E&F) Tg(sox10:memRFP) hmgcs1 siblings or homozygous hmgcs1 mutant larvae were imaged for RFP expression at 3dpf at the protruding mouth stage. Numbers indicate structures that will give rise to the putative ceratobranchial cartilages. n = 20 including 14/20 wildtype and heterozygous siblings and 6/20 homozygous mutants. Panel E depicts a wildtype sibling with no significant differences observed in heterozygous carriers. EXPRESSION / LABELING:
PHENOTYPE:
|
|
hmgcs1 regulates facial development by a cholesterol independent mechanism. (A-C) Wildtype AB embryos were treated with 0.01% DMSO (vehicle), 8uM lonafarnib (Lona) or 3 uM Ro 48 8071 (Ro-48) beginning at 5 hours post fertilization and for a period of 4 days post fertilization (dpf). Alcian blue was used to analyze the cartilage structures of the developing zebrafish head and neck. (A-C) demonstrate the dissected viscerocranium of 4 day old larvae. (DMSO n = 30, Ro-48 n = 14, and Lona n = 8) (D-F) Whole mount in situ hybridization (ISH) was performed at 3 days post fertilization (protruding mouth stage) with embryos treated with 0.01% DMSO (vehicle), 8uM lonafarnib (Lona) or 2.5 uM Ro 48 8071 (Ro-48) (DMSO n = 20, Ro 48 n = 27, and Lona n = 30) (G) Schematic representation of viscerocranium. The extension of the Meckel’s cartilage (mandible) was measured as a detection for the presence of a facial cleft phenotype. Distance is indicated by the red line. (G’) Average was normalized and represented relative to the vehicle control group. Asterisk denotes statistical significance of p<0.001. Statistical analysis was performed using a T-test. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Inhibition of isoprenoids and cholesterol has not effect on sonic hedgehog (Shh) activity. (A-C) Whole mount in situ hybridization (ISH) was performed at 3 days post fertilization (protruding mouth stage) (dpf) with embryos treated with 0.01% DMSO (vehicle), 8uM lonafarnib (Lona) or 2.5 uM Ro 48 8071 (Ro-48). (DMSO n = 10, Ro-48 n = 13, and Lona n = 16). (D) Sybr green based quantitative real time PCR was performed on hmgcs1 mutant embryos (Homozygous MT) or their wildtype siblings (WT Sibling) at the indicated days post fertilization (DPF) to detect mRNA expression of gli1. Error bars represent standard deviation of technical replicates. EXPRESSION / LABELING:
|
|
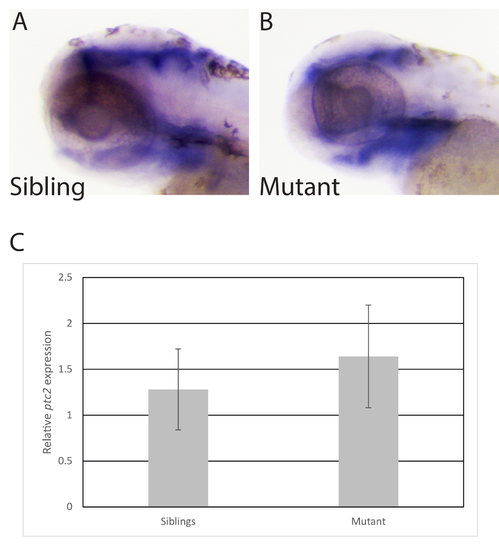
Mutations in hmgcs1 do not affect sonic hedgehog signaling. A-B. Whole mount in situ hybridization was performed on wildtype and mutant siblings at 3 days post fertilization/protruding mouth stage to detect the expression of ptch2. n = 47 including 17 wildtype, 7 homozygous mutants, and 23 heterozygous mutants. C. Real time PCR was performed on a pool (n = 20) of siblings or hmgcs1 mutant embryos at 4 days post fertilization to detect ptch2 expression (ptc2). Error bars demonstrate the standard error of the mean across two biological replicates each with 20 embryos per pool. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|