- Title
-
Yap and Taz regulate retinal pigment epithelial cell fate
- Authors
- Miesfeld, J.B., Gestri, G., Clark, B.S., Flinn, M.A., Poole, R.J., Bader, J.R., Besharse, J.C., Wilson, S.W., Link, B.A.
- Source
- Full text @ Development
|
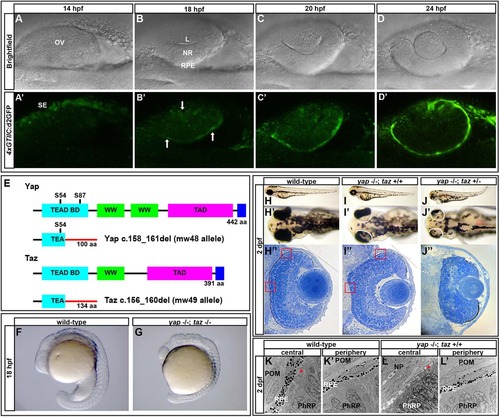
A Yap/Taz-Tead reporter transgene is dynamically expressed during optic cup morphogenesis and yap-/- mutants exhibit RPE defects. (A-D′) Images of live zebrafish from 14-24hpf showing optic cup development and 4xGTIIC:d2GFP transgene expression (green). Arrows indicate cells that are expressing the transgene while undergoing morphogenesis. (E) Schematic of wild-type and mutant Yap and Taz. Yap S54 is an essential residue for Tead binding and S87 is phosphorylated by Lats leading to cytoplasmic retention. The Yap c.158_161del and Taz c.156_160del mutants contain frameshifts resulting in early stop codons. TEAD BD, Tead transcription factor binding domain (light blue); WW, dual tryptophan motif (green); TAD, transactivation domain (fushia); PDZ domain (dark blue). (F,G) yap-/-; taz-/- embryos arrest by 18hpf with multiple defects. (H-J′′) Live embryos (H-J′) and sections (H′′,I′′,J′′) of yap-/-; taz+/+ (I-I′′) and yap-/-; taz+/- (J-J′′) showing RPE defects and additional NR defects in yap-/-; taz+/- mutants (J′) compared with control (H-H′′). Boxed areas indicate locations of TEM analysis. (K-L′) Transmission electron microscopy analysis showing areas of normal RPE development (L′) and areas devoid of RPE (L) in yap-/- eyes. Asterisk indicates the presence of primary cilia on neuroepithelial cells. L, lens; OV, optic vesicle; NR, neural retina; RPE, retinal pigment epithelium; SE, surface ectoderm; POM, periocular mesenchyme; NP, neuropil; PhRP, photoreceptor progenitors. EXPRESSION / LABELING:
PHENOTYPE:
|
|
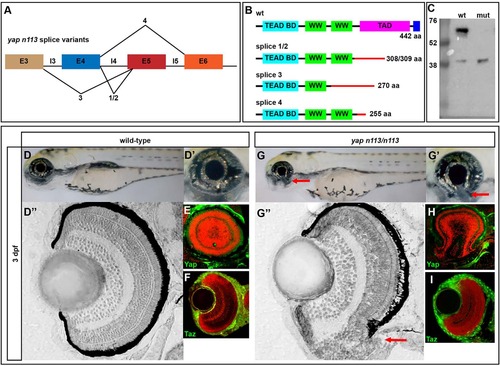
yapnl13/nl13 mutants exhibit coloboma. (A) Splice site variants elicited by the yapnl13 allele. The mutation results in aberrant splicing and an early stop codon in the transactivation domain for all described products. (B) Protein schematics for wild-type and predicted mutant yapnl13 allele variants. (C) Western blot showing the loss of full-length Yap in yapnl13/nl13 mutants. A smaller protein product is detectable at higher levels in the mutant (~40 kDa). (D-D′′,G-G′′) 3dpf wild-type and yapnl13/nl13 embryos showing coloboma (arrows) in the absence of other overt phenotypes. Plastic sections of wild-type (D′′) and mutant (G′′) eyes show the coloboma (arrow) phenotype and RPE deficits that are sometimes observed in the ventral retina. (E,F,H,I) Sections showing Yap and Taz proteins (green) in wild-type and yapnl13/nl13 mutant eyes. Red counterstain (TO-PRO3) shows nuclei. EXPRESSION / LABELING:
PHENOTYPE:
|
|
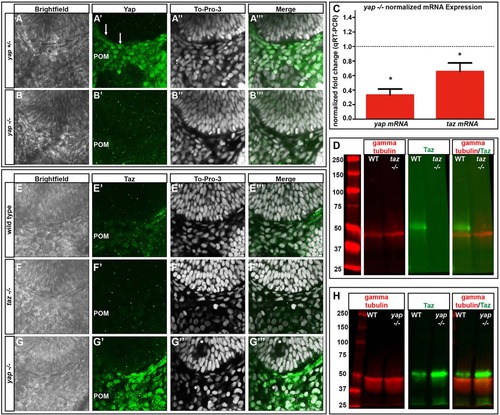
yap mRNA and Yap protein levels are decreased and Taz protein increased in yap-/- embryos. (A-B′′′) Yap immunoreactivity in wild-type and yap-/- eyes at 28hpf. Yap protein is present in flattened RPE nuclei (arrows) and periocular mesenchyme (POM) in yap+/- embryos, whereas nuclear Yap staining is absent in the yap-/- mutant. (C) qRT-PCR analysis of whole embryos at 32hpf showing a decrease in yap (3-fold, *P=0.0002) and taz (1.5-fold, *P=0.0270) mRNA in yap-/- mutants. Dotted line indicates normalized expression levels of yap and taz in wild-type embryos. An unpaired t-test was performed and statistical significance determined using the Holm-Sidak method. Error bars represent s.e.m. (D) Western blot showing Taz protein (~52 kDa) in wild-type and its absence in taz-/- adult heart tissue. (E-G′′′) Taz immunoreactivity in wild-type, taz-/- and yap-/- embryos at 28hpf. (H) Western blot of Taz protein from 2dpf wild-type (n=20) and yap-/- mutant (n=20) whole embryos. |
|
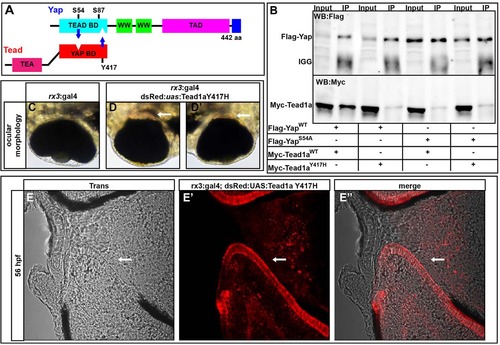
Yap and Tead1a zebrafish protein interactions are conserved. (A) Schematic of the zebrafish Yap and Tead binding domain (BD) interaction sites. (B) Immunoprecipitation of zebrafish Yap and Tead1a wild-type and Tead-binding-deficient Yap (Yap S54A) and Yap-binding-deficient Tead1a (Tead1a Y417H) isoforms. All the mutated protein variants lose the ability to interact, in contrast to the wild-type proteins. Immunoprecipitation (IP) was with an anti-Flag antibody. (C-E′′) Whole eyes (C-D′) and sections (E-E′′) showing RPE loss surrounding the optic nerve head after overexpression of Tead1a Y417H. Photoreceptors are red owing to late expression of the rx3:Gal4 driver in these cells. Arrows indicate areas lacking RPE. |
|
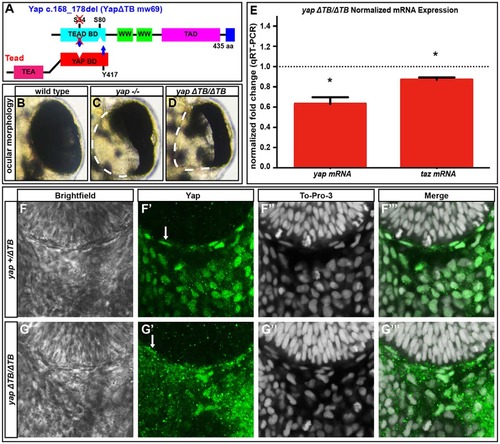
Tead-binding-deficient yapΔTB/ΔTB mutants lack RPE but maintain yap mRNA and Yap protein levels. (A) The Tead-binding-deficient yapΔTB/ΔTB zebrafish mutant. (B-D) 48hpf whole eyes showing that yap-/- and yapΔTB/ΔTB mutants lose a subset of RPE cells. Dashed lines indicate the border of the eye. (E) qRT-PCR analysis of whole embryos at 32hpf revealing a decrease in yap (1.6-fold, *P=0.0052) and taz (1.15-fold, *P=0.0038) mRNA in yapΔTB/ΔTB mutants. Dotted line indicates the normalized expression levels of yap and taz in wild-type embryos. An unpaired t-test was performed and statistical significance was determined using the Holm-Sidak method. Error bars indicate s.e.m. (F-G′′′) Yap protein expression in yap+/ΔTB (F-F′′′) and yapΔTB/ΔTB (G-G′′′) at 28hpf. Yap protein is present in flattened RPE nuclei (arrows) and periocular cells in yap+/ΔTB and yapΔTB/ΔTB embryos. |
|
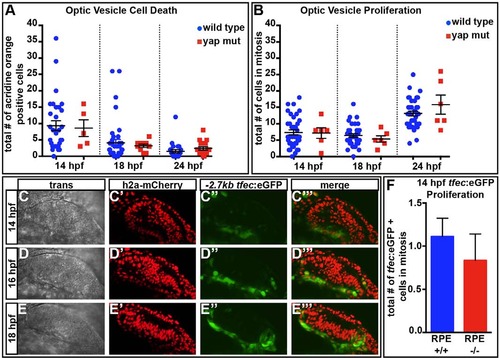
Cell death and proliferation are normal in yap-/- eyes. (A) The numbers of dying cells identified by Acridine Orange staining do not differ in yap-/- mutants at 14 (P=0.8465), 18 (P=0.6542) or 24 (P=0.2558) hpf as compared with wild type. Numbers of eyes analyzed: 14hpf, wt n=31, yap-/- n=5; 18hpf, wt n=39, yap-/- n=10; 24hpf, wt n=24, yap-/- n=16. (B) Eye field mitotic cell counts do not differ between yap+/? (wt) and yap-/- at 14 (P=0.9205), 18 (P=0.4329) and 24 (P=0.2222) hpf. Numbers of eyes analyzed: 14hpf, wt n=38, yap-/- n=6; 18hpf, wt n=38, yap-/- n=6; 24hpf, wt n=36, yap-/- n=6. (C-E′′′) Time-course of expression of tfec:eGFP in prospective RPE cells. (F) Mitotic cell counts of tfec:eGFP+ cells do not differ between wild-type and yap-/- eyes at 14hpf (P=0.5408). Numbers of eyes analyzed: wt n=18, yap-/- n=6. P-values were obtained using an unpaired t-test with equal s.d. Error bars indicate s.e.m. Wild type included full RPE coverage, whereas yap-/- showed some RPE loss. EXPRESSION / LABELING:
|
|
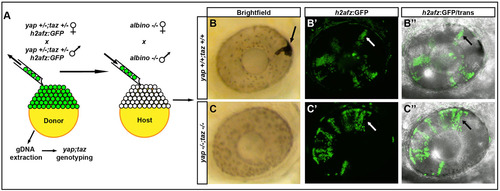
yap-/-; taz-/- transplanted cells do not contribute to the RPE. (A) Methods used to analyze the contribution of transplanted cells to the RPE and NR. (B-C′′) Examples of transplanted wild-type (B-B′′) and yap-/-; taz-/- (C-C′′) cells in 48hpf eyes. The arrow in B indicates a pigmented transplanted cell forming RPE in the albino host; other black/white arrows indicate H2a-GFP+ clones in NR. |
|
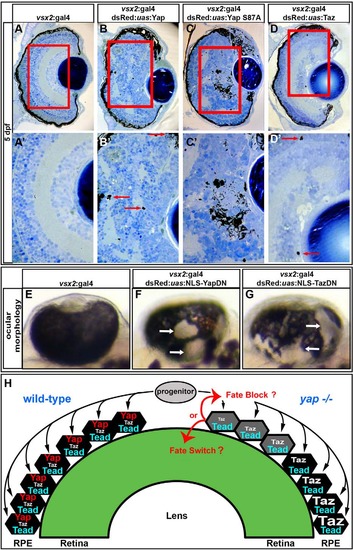
Overexpression of gain- and loss-of-function yap transgenes alters RPE and NR cell fate. (A-D′) Images showing that overexpression of wild-type Yap (B,B′), constitutively active Yap (Yap S87A) (C,C′) and Taz (D,D′) induces ectopic pigmentation and retinal disorganization at 5dpf, as compared with the control (A,A′). Arrows indicate the presence of ectopic pigment cells. (E-G) Mosaic overexpression of dominant-negative forms of Yap (F) and Taz (G) results in ectopic loss of RPE cells at 48hpf, as compared with the control (E). Arrows indicate areas devoid of RPE cells. (H) Model of zebrafish RPE development. Bipotent progenitor cells assume either an RPE or NR fate based on Yap/Taz activity. In the absence of yap (right hemisphere of the eye cup), progenitors contributing to the central retina cannot form RPE. Those progenitors that contribute to the peripheral eye cup can upregulate Taz during their more lengthy migration and therefore form RPE. |
|
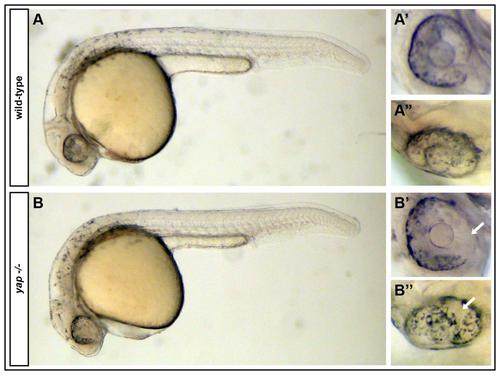
yap-/- mutants exhibit RPE defects noticeable at the onset of pigmentation. (A-A′′) A Wild-type embryo with normal RPE cells and pigmentation encompassing the whole eye globe. (B-B′′) yap-/- embryos lack RPE cells before RPE cells are completely pigmented. White arrows = areas of absent of RPE cells. |
|
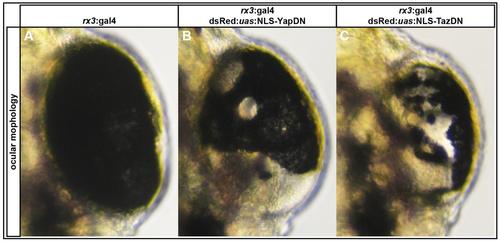
Overexpression of NLS-YapDN and NLS-TazDN in rx3 positive cells results in loss of RPE. (A-C) Examples of eyes from rx3:gal4+/dsRed:uas:NLS-YapDN and rx3:gal4+/dsRed:uas:NLS-TazDN fry lacking RPE cells at 48 hpf. |
|
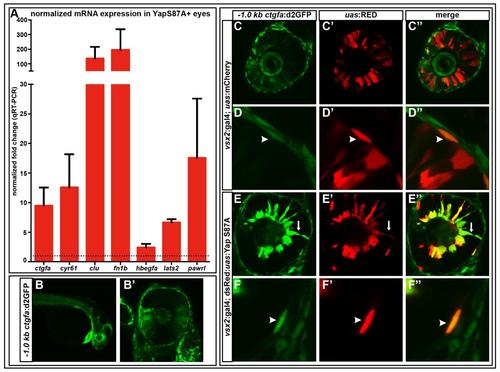
qRT-PCR validates mRNA transcripts that were upregulated via RNAseq in Yap S87A overexpressing 36 hpf eyes and -1.0kb ctgfa:d2GFP is upregulated by Yap S87A. (A) ctgfa (9.5-fold, p=0.0508), cyr61 (12.6-fold, p=0.1072), clu (135.5-fold, p=0.1705), fn1b (195-fold, p=0.2418), hbegfa (2.4 –fold, p=0.0845), lats2 (6.7-fold, p=0.0005), pawrl (17.6-fold, p=0.1736). Dashed line indicates the normalized expression levels of yap and taz in wild-type embryos. An unpaired t-test was performed on mRNA expression and statistical significance was performed using the Holm-Sidak method. Error bars = s.e.m. (B-B′) -1.0kb ctgfa:d2GFP expression is observed in the NR, RPE, heart, and other tissues at 28 hpf. (C-F′′) Yap S87A overexpression results in increased -1.0kb ctgfa:d2GFP in RPE cells (F-F′′) and ectopic NR expression (E-E′′) compared to mCherry (C-D′′). Arrows=ectopic NR, arrow heads=RPE expression. All embryos are 28 hpf. |
|
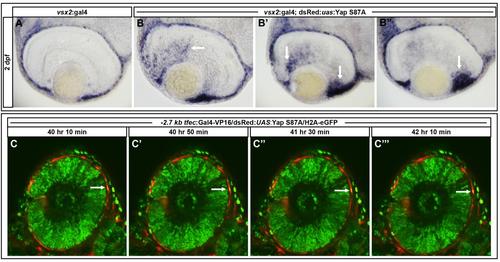
Overexpression of Yap S87A in vsx2 positive cells results in ectopic expression of dct mRNA and -2.7kb tfec:Gal4-VP16/dsRed:UAS:Yap S87A positive cells do not migrate into the neural retina. (A) Endogenous dct expression is observed in the RPE. (B-B′′) Ectopic dct expression is observed in the neural retina and enhanced in the ciliary marginal zone in Yap S87A overexpressing embryos. Black arrows denote ectopic dct. (C-C′′′) RPE cells overexpressing Yap S87A do not ectopically migrate into the neural retina. Images represent a 2 hour time course from 40-42 hpf. White arrows indicate a Yap S87A positive RPE cell. |